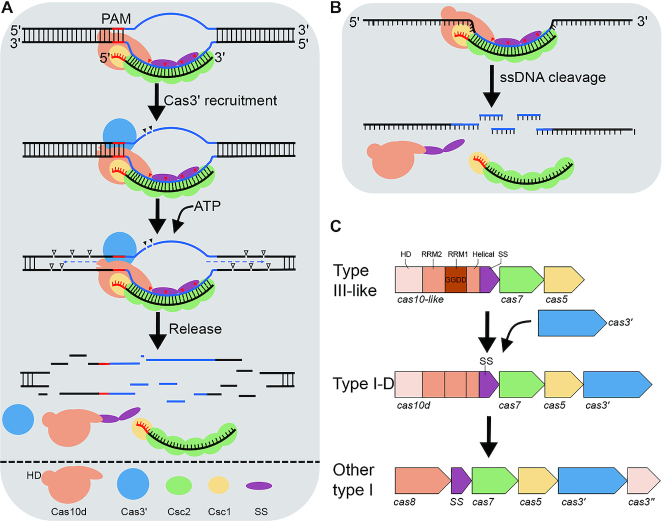
Figure 4.
Interference mechanisms and evolution of type I-D CRISPR–Cas system. (A) dsDNA interference by the type I-D effector. Cas10d recongnizes PAM, enabling base-pairing between guide sequence of crRNA and the target strand of protospacer, forming an R-loop. The Cas3′ helicase is recruited, Cas10d nicks the non-target strand of the protospacer. Next, Cas3′ unwinds the DNA at both directions of the protospcer, and Cas10d cuts ssDNA of both strands at multiple sites. Possibly, release of the cleavage products shuts off Cas10d activity, completing the DNA interference. Since neither Cas10d nor SS were detected in the backbone complex purified from Sulfolobus (Figure 1B), we infer that Cas10d/SS do not form a stable complex with the backbone in the absence of a target dsDNA. (B) Ruler-based ssDNA interference. The type I-D effector cleaves ssDNA targets at 6 nt intervals within the protospacer region, and no PAM is required. (C) Evolution model of type I CRISPR–Cas systems from a type III-like ancient system via a type I-D like intermediate. RRM: RNA recognition motif, SS: small subunit.