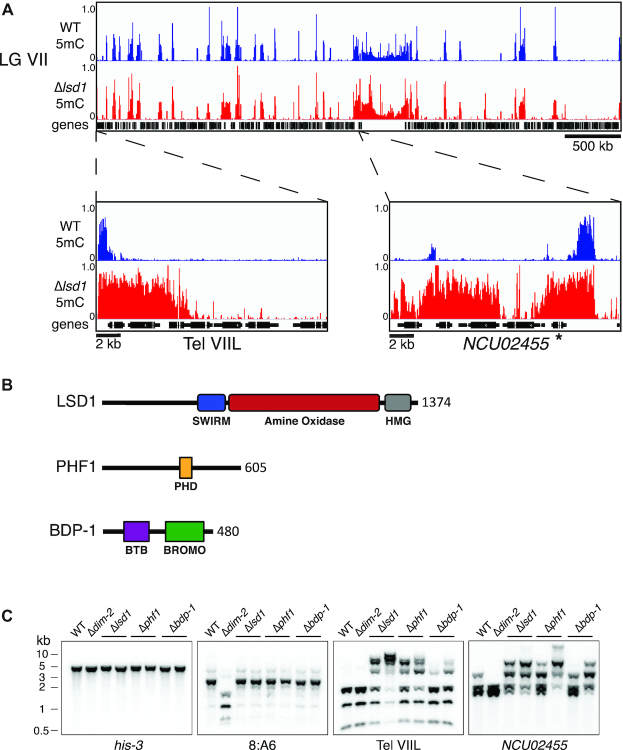
Figure 1.
Identification of LSDC in Neurospora crassa. (A) WGBS tracks displaying DNA methylation in WT (blue) and Δlsd1 (red) strains over LG VIIL (additional tracks for other chromosomes can be found in Supplemental Figure S2). NCU02455 is indicated with an asterisk. (B) Schematic representation of the identified members of LSDC with their predicted domains and length (amino acids) indicated. (C) LSDC knockouts exhibit hypermethylation. Genomic DNA from WT, a strain lacking DNA methylation (Δdim-2), and LSDC knockouts (Δlsd1, Δphf1 and Δbdp-1) were digested with the 5mC-sensitive restriction enzyme AvaII and used for Southern hybridizations with the indicated probes for a euchromatin control (his-3), an unaffected heterochromatin control (8:A6), and two Δlsd1-sensitive regions (Tel VIIL and NCU02455). Strains (left to right): N3753, N4711, N5555, N6411, N6221, N6414, N6220 and N6416.