Figure 3.
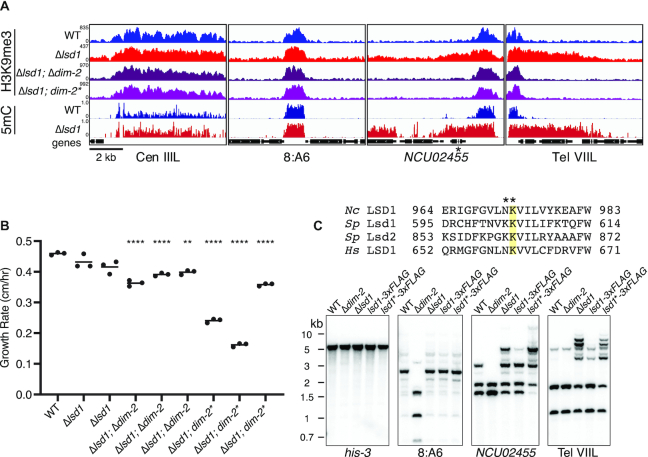
LSD1 prevents DNA methylation-dependent hyper H3K9me3. (A) H3K9me3 ChIP-seq and WGBS tracks showing H3K9me3 enrichment and DNA methylation respectively at two unaffected heterochromatin regions (Cen IIIL and 8:A6) and two Δlsd1-sensitive regions (NCU02455 and Tel VIIL) in the indicated strains. dim-2* denotes the C926A catalytic null mutation in dim-2 (42). NCU02455 is indicated with an asterisk. (B) Linear growth rates in WT, Δlsd1, and Δlsd1 strains bearing a deletion (Δ) or catalytic null (*) mutation in dim-2. Strains (left to right): N3753, N5555, N7979, N6337, N8081, N8082, N6679, N8083 and N8084. All Δlsd1; Δdim-2 and Δlsd1; dim-2* strains are siblings from separate crosses. (****) P ≤ 0.0001; (**) P ≤ 0.01. (C) Loss of LSD1 catalytic activity causes hyper DNA methylation. (Top panel) Sequence alignments of human, S. pombe, and Neurospora LSD1 homologs centered on a lysine essential for catalytic activity (highlighted). The NK982,983AA mutation introduced to create the presumptive catalytic null lsd1 Neurospora strains is displayed above. (Bottom panel) DNA methylation-sensitive Southern hybridization analysis (as in Figure 1C) on 3xFLAG-tagged and catalytic null (indicated by *) 3xFLAG-tagged LSD1 strains. Strains (left to right): N3753, N4711, N5555, N6300 and N7899.