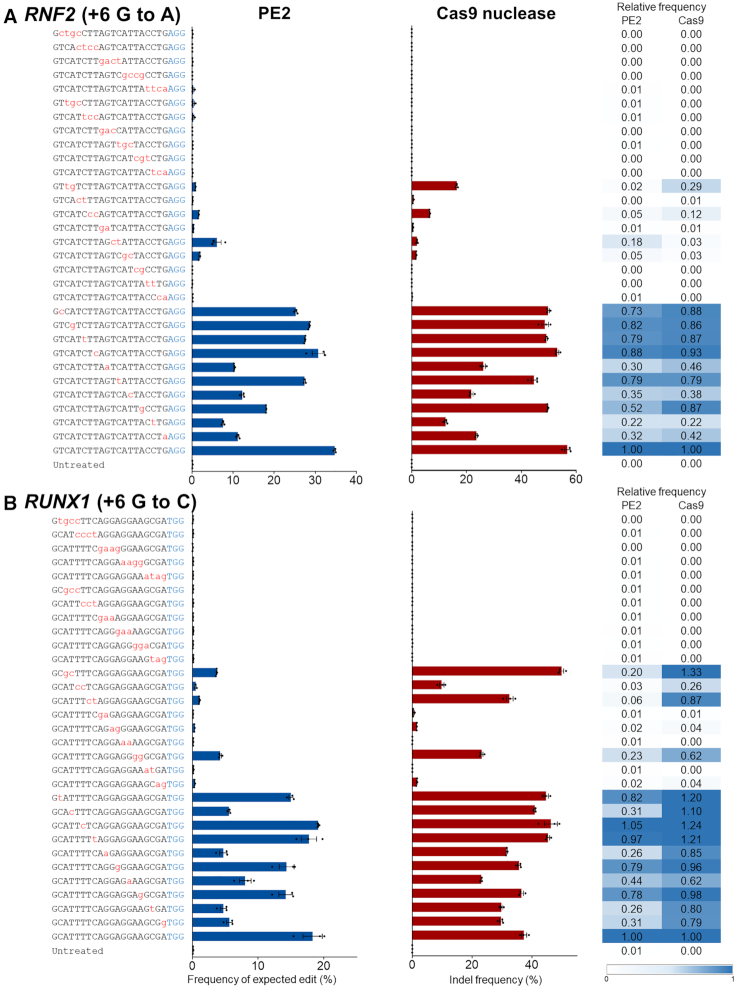
Figure 1.
Tolerance of PE2 and Cas9 nuclease for mismatched pegRNAs or sgRNAs, respectively. (A and B) Together with plasmids encoding PE2 or Cas9 nuclease, plasmids encoding pegRNA or sgRNA that differed from the RNF2 (A) and RUNX1 (B) target sequences by 1–4 nt were respectively transfected into HEK293T cells. Editing frequencies and indel frequencies were measured by targeted deep sequencing. The PAM is shown in blue and the mismatched nucleotides are shown in red lowercase letters. Error bars indicate the s.e.m. (three biologically independent samples). Relative frequencies were calculated by dividing the frequency of edits or indels obtained using mismatched pegRNAs or sgRNAs by the frequency of the on-target edits.