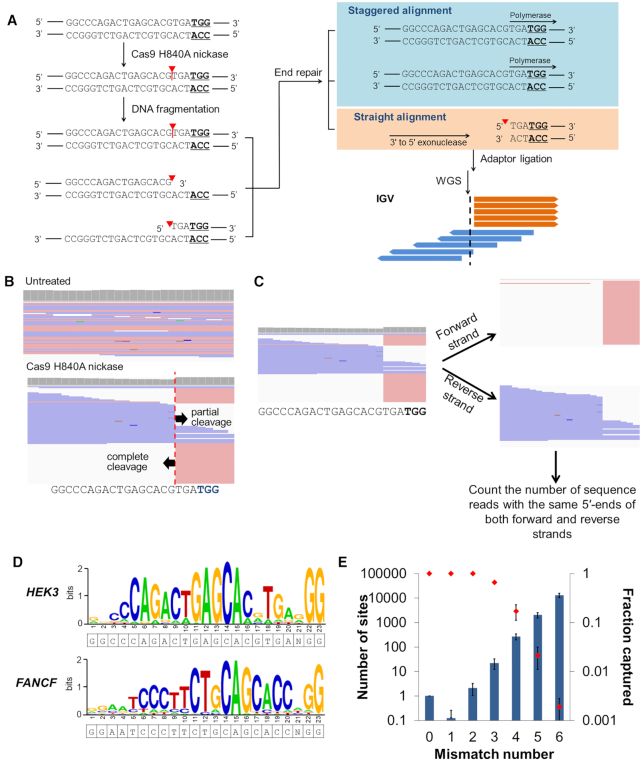
Figure 2.
nDigenome-seq to identify SSB sites generated in vitro in the human genome by Cas9 H840A. (A) Overview of nDigenome-seq. gDNA is prepared from cells and treated with Cas9 H840A nickase and sgRNA, resulting in a nick in the protospacer of on-target and off-target sites. The fragmentation step, done by sonication, renders a library of ∼500 bp fragments with random ends. Treatment with a polymerase with 3′ to 5′ exonuclease activity leads to end repair, during which the 3′-end resulting from the nick (that is, fragments with a 5′-overhang) is filled. As a result, the filled strand shows a random end sequence due to the random fragmentation, thereby exhibiting staggered alignment in an IGV file. However, the fragments with a 3′-overhang are trimmed by 3′ to 5′ exonuclease activity, resulting in fragments with blunt ends. Thus, the trimmed fragments show a straight alignment pattern in an IGV file. In summary, a straight alignment pattern along with a staggered alignment pattern indicates the occurrence of a nick induced by Cas9 H840A nickase at both on-target and off-target sites. We used a TruSeq DNA Library Prep Kit (illumina) for adaptor ligation and to generate a sequencing library. Forward and reverse sequence reads are shown in pink and blue, respectively. Red triangles and vertical lines represent cleavage positions. (B) An IGV image of sequence reads that reveals complete and partial cleavage of the non-target and target strands, respectively, at the HEK3 on-target site. (C) Aligned sequence data were separated into forward and reverse strands, and the number of sequence reads with the same 5′ end at each position were counted to identify in vitro SSB sites. (D) Representative sequence logos for the HEK3 and FANCF sites were obtained from WebLogo using DNA sequences at the sites captured by nDigenome (sequence read counts starting at this position ≥10 and count/depth ≥20%). (E) The number of potential off-target sites with homologous sequences (left y-axis; bars) increased exponentially as the number of mismatches with the nine Cas9 H840A on-target sites tested increased. In contrast, the fractions of captured sites (right y-axis, red dots) decreased sharply. Error bars indicate the s.e.m (n = 9).