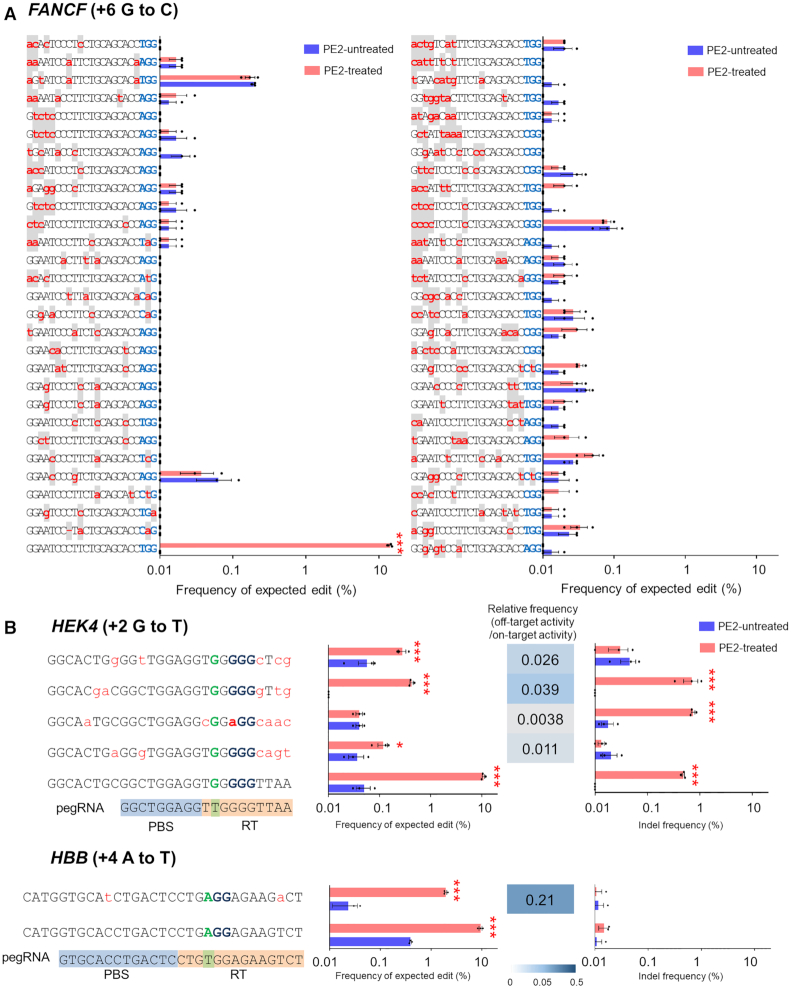
Figure 3.
Validation of PE2 off-target sites identified by Cas9 H840A nickase-mediated nDigenome-seq. (A) In vitro SSB sites captured by nDigenome-seq after treatment with Cas9 H840A nickase targeting FANCF (installing +6 G to C mutation) were subjected to validation by targeted deep sequencing using gDNA extracted from HEK293T cells transfected with plasmids encoding PE2 and pegRNA. The mismatched nucleotides are shown in red lowercase letters on a gray background, and PAM sequences are shown in blue. Error bars indicate the s.e.m. (three biologically independent samples). (B) On-target and validated off-target sites identified in human cells using targeted deep sequencing. The mismatched nucleotides are shown in red lowercase letters and PAM sequences are shown in blue. Precise edit and indel frequencies at the validated off-target sites for Cas9 H840A nickase targeted to HEK4 and HBB. PBS and RT template sequences are highlighted with blue and orange backgrounds, respectively. Error bars indicate the s.e.m. (three biologically independent samples). P-values were calculated by Student's t-test between PE2-untreated and PE2-treated. P -value, *; <0.05, **; <0.01, ***; <0.001.