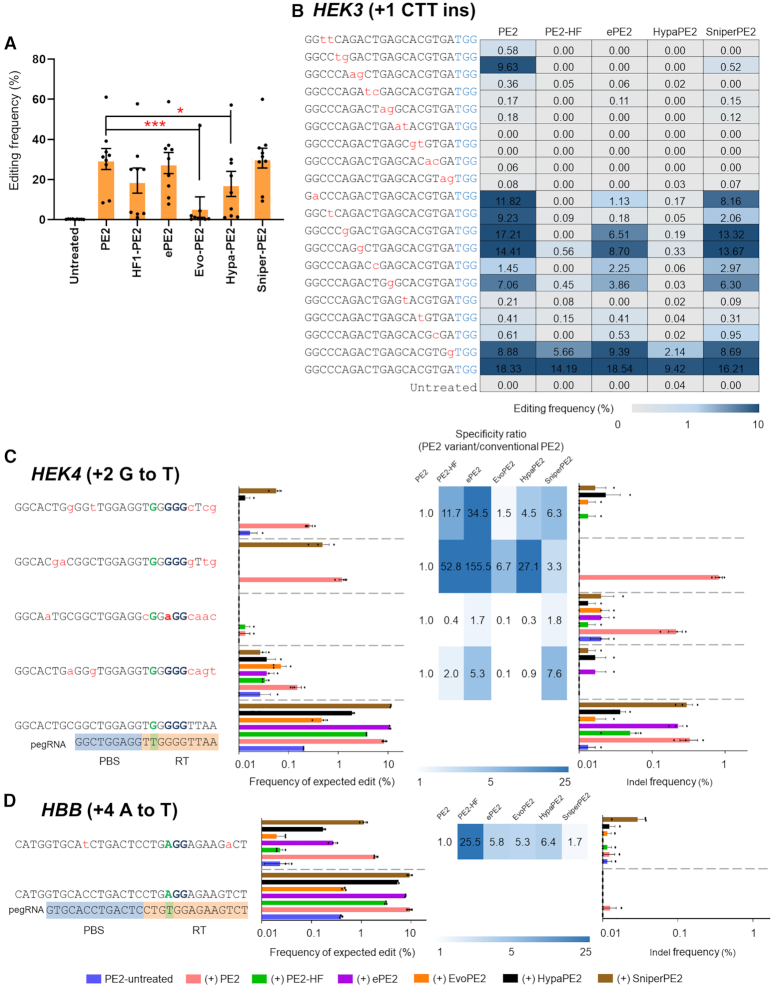
Figure 6.
Enhancing PE2 specificities with PE2 variants. (A) Precise edit frequencies achieved by PE2, PE2-HF, ePE2, Evo-PE2, Hypa-PE2 and Sniper-PE2 at nine selected endogenous target sites in HEK293T cells. Error bars indicate the s.e.m. (n = 9). P -values were calculated by Student's t-test. P-value, *; <0.05, **; <0.01, ***; <0.001. (B) Plasmids encoding mismatched pegRNAs that differed from the HEK3 site (installing +1 CTT ins) by one to 2 nt were transfected into HEK293T cells. Precise editing frequencies were measured by targeted deep sequencing. (C andD) Improved specificity of PE2 variants as validated at off-target sites with a high sequence similarity with the HEK4 (C) and HBB (D) on-target sites. Specificity ratios were calculated by dividing the specificity of conventional PE2 (on-target frequency/off-target frequency) by the specificity of the PE2 variant (on-target frequency/off-target frequency). The heatmap represents the relative specificity of the PE2 variants compared to that of the canonical PE2. The mismatched nucleotides are shown in red lowercase letters, and the PAM sequences are shown in blue. PBS and RT template sequences are highlighted with blue and orange backgrounds, respectively. Error bars indicate the s.e.m. (three biologically independent samples).