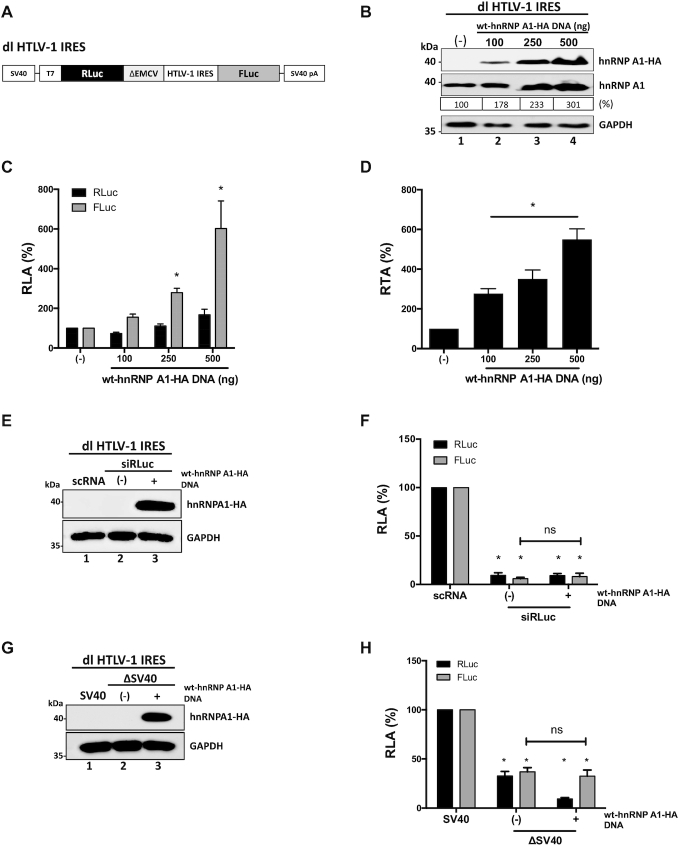
Figure 6.
HnRNP A1 is an ITAF for the HTLV-1 IRES. (A) Schematic representation of the dl HTLV-1 IRES plasmid described in (12). (B–D) HEK 293T cells were co-transfected with the dl HTLV-1 IRES (300 ng) and increasing amounts of the wt-hnRNP A1-HA (100, 250, 500 ng) plasmid. Total protein extracts were prepared 24 h post-transfection. (B) Western blots were performed to follow the expression level of total hnRNP A1 and wt-hnRNP A-HA proteins using GAPDH as a loading control. The relative level of hnRNP A1 protein (%) was estimated based on the intensity of immunoreactive bands by imageJ. (C, D) RLuc and FLuc activities were measured, and data are presented as RLA (C) or RTA (D). The RLA and RTA values obtained when in the absence (–) of transfected wt-hnRNP A1-HA were set to 100%. Statistical analysis was performed by an ordinary two-way (C) or one-way (D) ANOVA test (*P <0.05). (E, F) The dl HTLV-1 IRES (150 ng) was co-transfected with a control scRNA (100 nM) or with siRLuc (100 nM), in the presence (+), or the absence (–), of the wt-hnRNP A1-HA (250 ng) plasmid. Total protein extracts were prepared 48 h post-transfection. (E) The expression of the hnRNP A1-HA recombinant protein was determined by western blot, using the GAPDH protein as a loading control. (F) RLuc and FLuc activities were measured and expressed relative to the values obtained with scRNA, set to 100% (RLA). (G, H) HEK 293T cells were transfected with either the dl HTLV-1 IRES (150 ng) or a promoterless ΔSV40-dl HTLV-1 IRES (150 ng) vector in the presence (+), or the absence (–), of the wt-hnRNP A1-HA (250 ng) plasmid. Total protein extracts were prepared 24 h post-transfection. (G) The expression of the hnRNP A1-HA recombinant protein was determined by western blot, using the GAPDH protein as a loading control. (H) RLuc and FLuc activities were measured, and results are expressed as RLA relative to the activities obtained from the dl HTLV-1 IRES plasmid transfected in the absence of the wt-hnRNP A1-HA, set to 100%. Values shown are the mean (±SEM) for three independent experiments, each performed in duplicate. Statistical analysis was performed by an ordinary two-way ANOVA test (*P <0.05; ns, not significant).