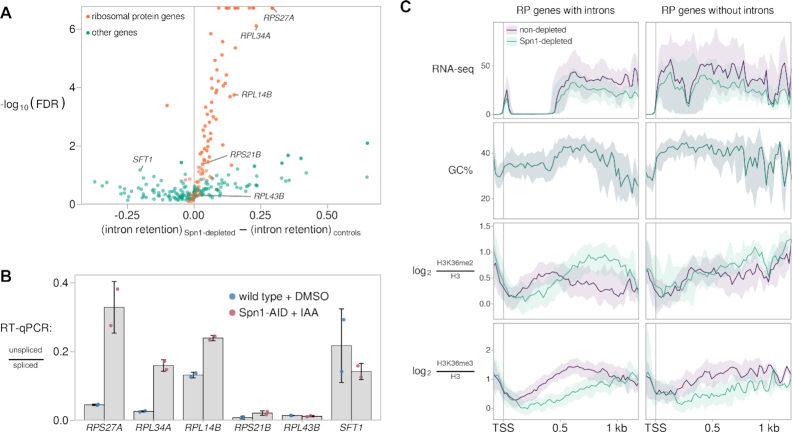
Figure 7.
Depletion of Spn1 results in increased intron retention in ribosomal protein transcripts. (A) Volcano plot showing changes in intron retention upon Spn1 depletion for 252 introns encoded from within coding regions of nuclear genes. Intron retention is defined as the proportion of unspliced transcripts and was estimated from RNA-seq data (see Materials and Methods). Labeled introns were further assayed by RT-qPCR, as shown in (B). (B) RT-qPCR measurements of the ratio of unspliced to spliced transcripts in a wild-type strain treated with DMSO for 90 min and the Spn1 degron strain treated with 25 μM IAA for 90 min. Error bars indicate the mean ± standard deviation of two replicates. (C) Visualization of average changes in H3K36 methylation state at ribosomal protein genes with and without introns. For each group of genes, the following data are plotted: sense strand RNA-seq coverage, GC% in a 21-bp window, and H3-normalized H3K36me2 and H3K36me3 ChIP enrichment. The solid line and shading are the median and interquartile range over the genes in each group. The introns of RP genes tend to be close to the 5′ end of the gene, the region where RNA-seq signal is seen as depleted (top left panel).