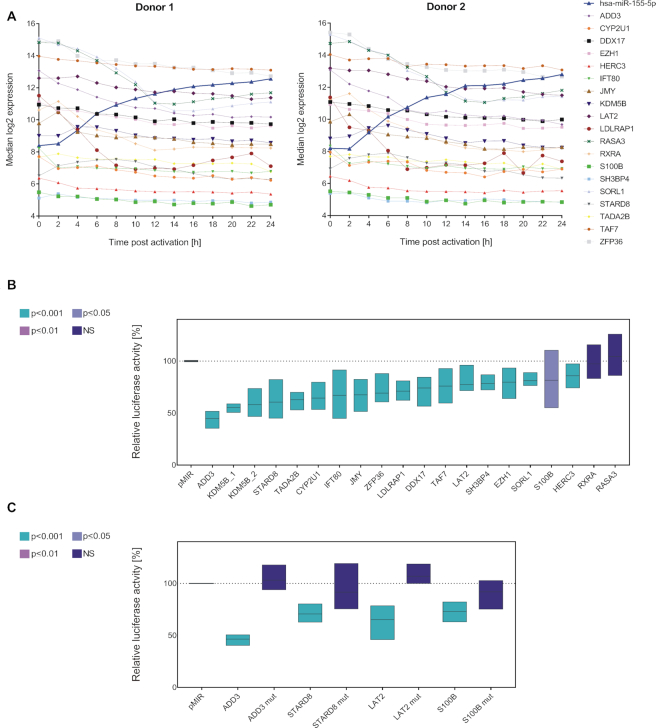
Figure 7.
MiR-155-5p and its target genes. (A) Overview on the time-resolved expression patterns of miR-155-5p and of its potential target genes (mRNA). Time courses (log2 expression) are donor-wise represented as median result of the three activation reactions per time point. Putative targets were selected based on an in silico prediction and an inverse correlation between their time-resolved mRNA profile and the corresponding miR-155-5p expression. (B) The interactions of miR-155-5p with its putative target genes were analyzed by dual luciferase assays. 3′UTR sequences of putative target genes were cloned into luciferase reporter plasmids (pMIR-RNL-TK) and tested in presence of miR-155 expression plasmid (pSG5-miR-155) or (empty) pSG5 control, respectively. The KDM5B 3′UTR was subdivided into two reporter constructs. Firefly and Renilla luciferase activities were measured 48 h after transfection of HEK293T cells. Results were standardized based on the transfection efficiency (determined by Renilla luciferase activity) and the basic activity of the 3′UTR-plasmid (empty pSG5 co-transfection). Results are shown in relation to the activity of an empty reporter control (pMIR without 3′UTR with miR-155 co-transfection ≡ 100%) as the average (line) with range (bars) of three independent experiments (conducted in duplicates). Statistical evaluation was performed in comparison to empty reporter control. P-values were adjusted by Benjamini–Hochberg. (C) For the exemplary validation of the luciferase assays, putative miR-155 binding sites in the 3′UTRs of four positively tested target genes were mutated and tested in comparative luciferase assays with both the wild type and corresponding mutated reporter constructs.