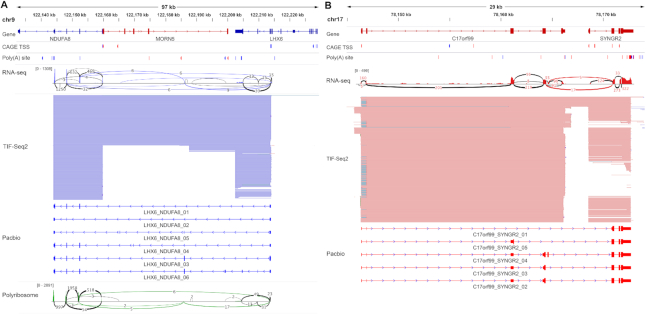
Figure 4.
TIF-Seq2 facilitates the identification of read-through transcripts. (A) LHX6-NDUFA8 read-through gene. (B) C17orf99-SYNGR2 read-through gene. Annotated genes are listed on the top, followed by CAGE TSS and poly(A) site (PAS) (12,13) tags which are represented by red or blue bars (forward or reverse strands respectively). RNA-Seq validates the presence of splicing junctions (red or blue lines with supporting number of reads) linking two adjacent genes. TIF-Seq2 track (as in Figure 2A) shows the transcriptional fusion events between adjacent genes. PacBio Long-read sequencing of target transcripts validates the intergenic splicing events and dissect the transcription model of read-through genes. Polyribosome-associated RNA-Seq (28) data are labelled in green, with splicing junctions in green lines between two genes, showing the coding potential of two transcript isoforms of LHX6-NDUFA8.