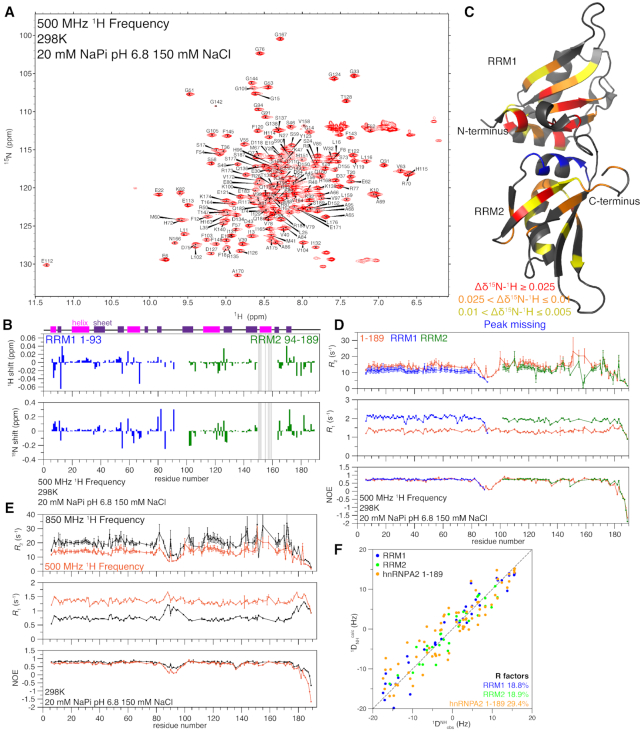
Figure 1.
hnRNPA2 RRMs remain folded and interact in the absence of RNA but do not form a rigid structure. See also Supplementary Figure S1. (A) 1H–15N HSQC of hnRNPA2 1–189 is consistent with a folded protein. (B) Chemical shift deviations for the individual RRMs compared to 1–189 are small and primarily at the N- and C-termini of the constructs (e.g. due only to primary structure differences), except in RRM2 where shifts were too large to determine peak identity by overlay, as indicated by grey bars. Secondary structure elements as determined by NMR (see Supplementary Figure S1A) shown at top, α-helices in magenta, β-sheets in purple. (C) Average chemical shift deviations (plotted in Supplementary Figure S1C) of difference between individual RRM spectra and tandem RRM spectra mapped on the structure of hnRNPA2 RRMs (PDB: 5HO4) to show where RRM1 and RRM2 interact with each other in the absence of RNA. (D) NMR spin relaxation parameters 15N R2, 15N R1, and hetNOE values for 1–189 (orange), RRM1 (1–93 blue), and RRM2 (94–189 green) at 500 MHz 1H frequency indicate the RRM move similarly but have slowed tumbling when the RRMs are linked. (E) NMR spin relaxation parameters 15N R2, 15N R1 and hetNOE values for 1–189 at 850 MHz 1H frequency (black) and 500 MHz 1H frequency (orange) indicate the presence of a flexible linker between the individual RRMs. (F) Agreement between the experimental (1DNHobs) RDCs and the back calculated (1DNHcalc) RDCs from PDB 5HO4 (15) for RRM1 (blue) and RRM2 (green) alone show lower R factors (better agreement) than for the tandem RRMs together (orange). See Table 2.