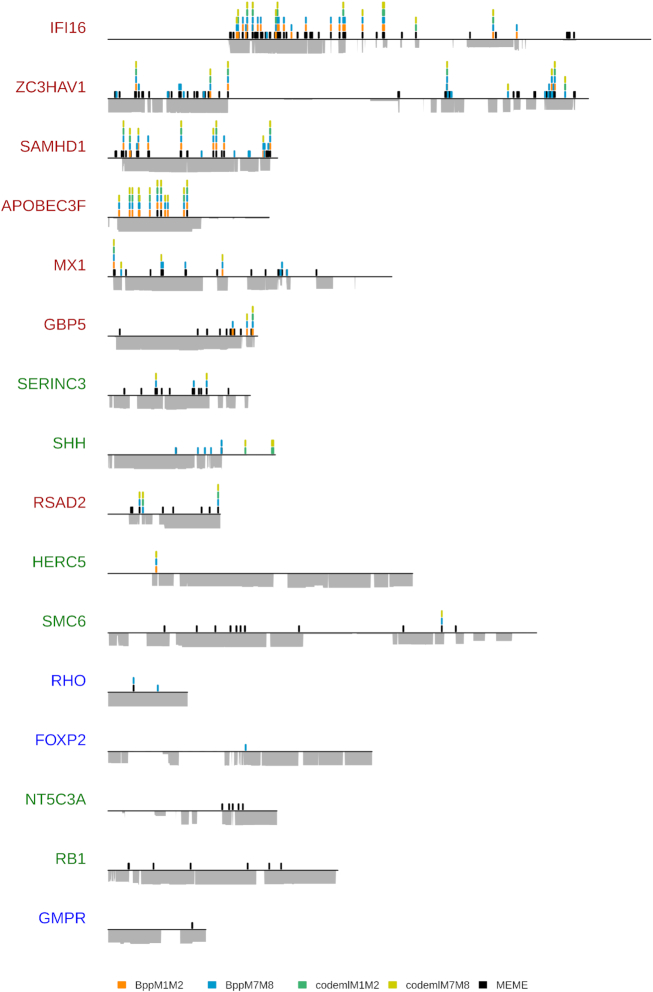
Figure 4.
Positive selection patterns on nineteen primate genes. The genes are color-coded according to their selection profile category (Table 2) and follow the same order as in Figure 3. Genes without positively selected sites were excluded from this representation. Positively selected sites are represented as a spike at their position on the alignment. Height of the peak is proportional to the number of methods that have identified the site as being under positive selection (posterior probabilities > 0.95 for Bio++ and PAML codeml M2 and M8 models, and P-value < 0.10 for MEME), with each method being represented by a different color (embedded legend). HYPHY MEME sites were only mapped if the gene was detected as under positive selection by BUSTED (P < 0.05). For each gene, alignment coverage is represented under the line, which itself represents the length of the alignment in light gray.