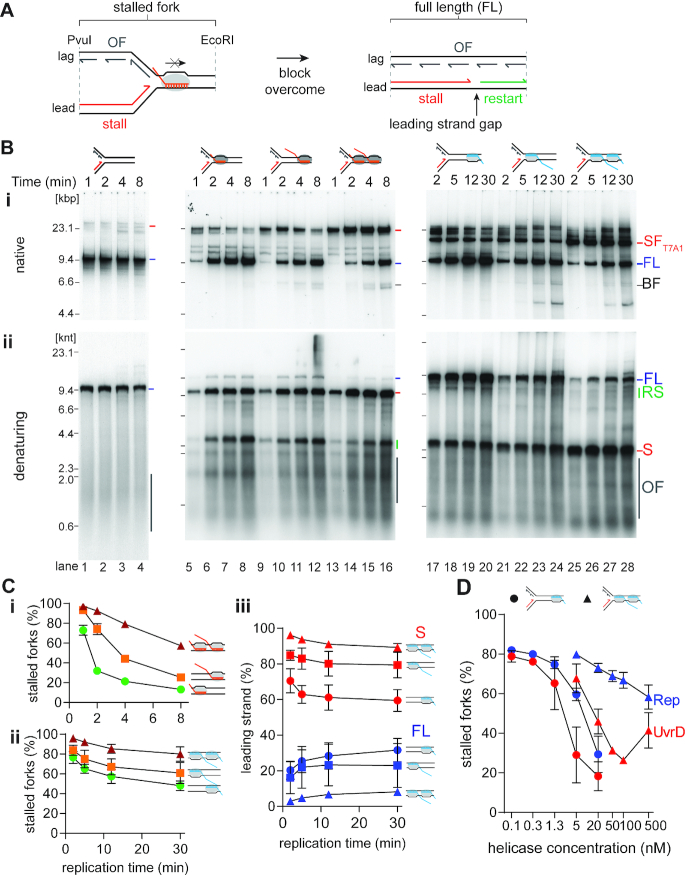
Figure 2.
Analysis of collisions between RNAP-template complexes and replisomes. (A) Cartoon illustrating the potential products of a collision between a replisome and a CO-RNAP-template complex. (B) CO RNAP-replication collisions cause transient fork stalling, whereas HO RNAP-replication collisions are severe blockages to fork progression. Time courses of replisome collisions on templates carrying either no RNAP complexes (on template CO100, lanes 1–4); three different CO RNAP-templates: 19mer (CO100), lanes 5–8, 100mer (CO100), lanes 9–12, or RNAP array (CO100), lanes 13–16; or three different HO RNAP-templates: 19mer (HO100), lanes 17–20, 100mer (HO100), lanes 21–24, or RNAP array (HO100), lanes 25–28, analyzed by either (i) native agarose gel or (ii) denaturing agarose gel electrophoresis (n = 5). (C) Fork blockage becomes more severe as the complexity of the transcription complex increases. The fraction of stalled fork DNA products from native gels are plotted as a function of time for CO (i) and HO (ii) collisions. (iii) Reciprocal relationship between the fraction of stalled leading strand product and full-length material as a product of time for HO collisions on denaturing gels. (D) Relative efficiency of the DNA helicases Rep (blue) and UvrD (red) in resolving stalled forks in HO collisions with a 19mer RNAP (•, on template HO19) or an RNAP array (▴, HO100) (n = 3). SF, stalled fork; FL, full length product; RS, restart product; S, stalled leading-strand product; OF, Okazaki fragments.