Figure 4.
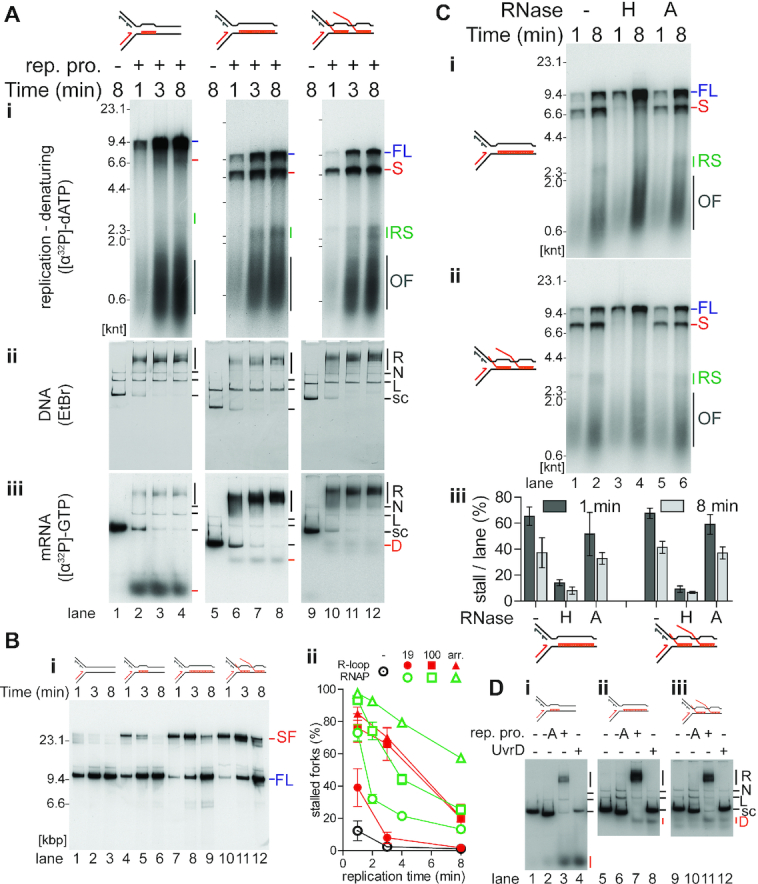
R-loops on the leading-strand template cause transient replisome stalling. (A) Time course of replisome collisions with three different R-loop species on the leading-strand template (n = 3). (i) Analysis of [α-32P]dATP-labeled replication reaction products on denaturing agarose gels. There is no DNA replication in the absence of the replication proteins as measured by the incorporation of [α-32P]dATP into acid-insoluble product. Thus, these samples, lanes 1, 5, and 9, were not included on the denaturing gel that is visualized by autoradiography. Native agarose gel of replication reaction products either stained with (ii) ethidium bromide or (iii) autoradiography of [α-32P]GTP-labeled RNA in the R-loops. Lanes 1–4, 19mer R-loop (on template CO19); lanes 5–8, 100mer R-loop (CO100); lanes 9–12, R-loop array (CO100). (B) Replication fork stalling increases with more complex R-loop species on the leading-strand template. (i) Native agarose gel of a replication time course of replisome collisions without or with leading-strand template R-loops. No R-loop (CO100), lanes 1–3; 19mer R-loop (CO19), lanes 4–6; 100mer R-loop (CO100), lanes 7–9; and R-loop array (CO100), lanes 10–12. Unlike the replication time courses in Figure 4A, these DNA templates had been linearized by ScaI digestion 40 s after replication initiation to allow for a more direct comparison between replication bypass of R-loops and CO replication-RNAP collisions. (ii) Quantification of stalled forks from native gels (n = 3, mean ± standard deviation) compared to the results from the quantification of stalled forks at CO replisome-RNAP collisions from Figure 2B lanes 5–16. R-loop collisions in red, RNAP collisions in open, green symbols. ⊙ no collision control, • 19mer, ▪ 100mer, ▴ array. (C) Replication stalling is relieved by treatment of the R-loop templates with RNase H, but not RNase A. Denaturing agarose gel of products in replication time courses of collisions with either the (i) CO 100mer R-loop template or (ii) CO R-loop array template subsequent to treatment with RNase H (lanes 3 and 4), RNase A (lanes 5 and 6), or no treatment (lanes 1 and 2). (iii) Quantification of the fraction of stall product (n = 3, mean ± standard deviation). (D) Either replication or UvrD treatment of R-loop templates can result in displacement of the RNA from the R-loop. Native agarose gel of [α-32P]GTP-labeled R-loop templates incubated (8 min) in replication buffer either without or with the indicated replication proteins and either with or without UvrD (100 nM), as indicated. (i) 19mer R-loop, (ii) 100mer R-loop, (iii) R-loop array. FL, full length leading strand; S, stalled leading strand product; RS, restarted leading strand products; OF, Okazaki fragments; R, replicated; N, nicked; L, linear; s.c., supercoiled; D, displaced RNA.
