Figure 4.
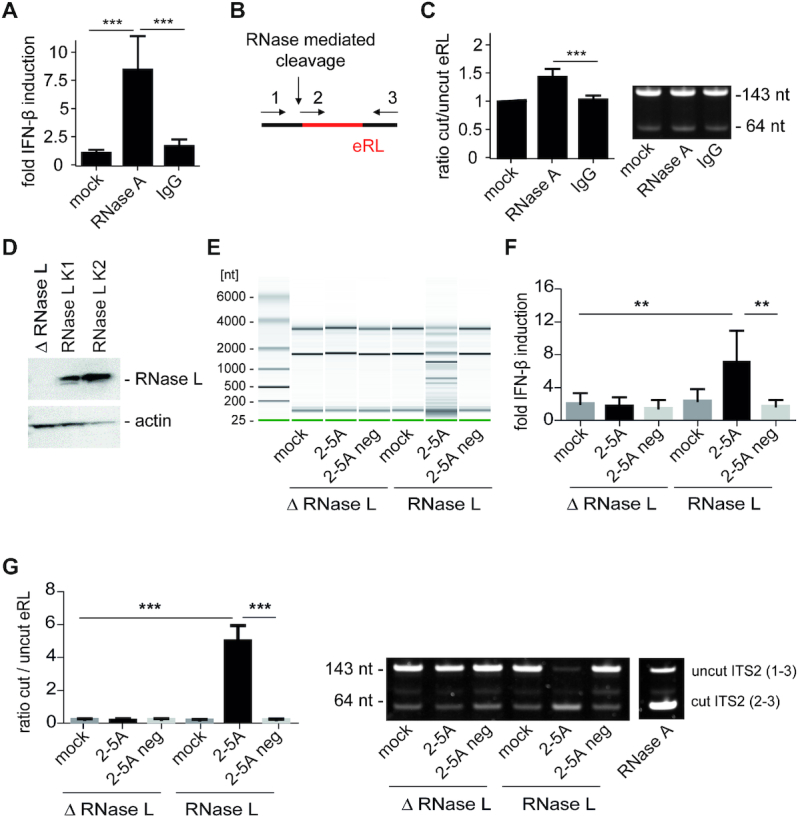
The eRL is generated intracellularly and induces immune activation. (A) HEK293-RIG-I-IFN-ß reporter cells were mock-treated or transfected with 3.5 μg/ml RNase A and 3.5 μg/ml murine IgG using SAINT-Protein transfection reagent and interferon reporter activity was measured 24 h after transfection. Graph combines three independent experiments each in biological quadruplicates (12 measurements per data point ± S.D.), *** P < 0.001. (B) Schematic illustration of the RT-PCR strategy using three primers (depicted as 1, 2 and 3, see Materials and Methods) in one reaction to analyze uncut or cut ITS2 fragments (eRL). (C) Cleavage of eRL was determined in qRT-PCR after 24 h. Left panel combines three independent experiments each in technical duplicates (six measurements per data point ± S.D). *** P < 0.001. Right panel depicts a representative fragment analysis in a 12% PAA gel stained with SYBRGold. One individual experiment of three independent experiments is shown. (D) Western blot of HEK293-RIG-I-IFN-ß reporter cells devoid of or reconstituted with RNase L. ß-actin served as loading control. (E) RNase L-deficient (△ RNase L) and RNase L-competent (RNase L K2) HEK cell lysates were untreated (mock) or incubated with 2′-5′-oligoadenylate (2–5A) containing lysate or control lysate (2–5A neg). After incubation of 1 h at 37°C, RNA was purified and RNA integrity investigated utilizing Pico RNA chips on an Agilent bioanalyzer. One individual representative experiment of three independent experiment is shown. (F) RNAs depicted in (E) were transfected into HEK293-RIG-I-IFN-β reporter cells at 5 μg/ml RNA each. Fold IFN-ß induction was determined after 12–16 h. Graph combines three independent experiments each in biological duplicates (6 measurements per data point ± S.D.), ** P < 0.01. (G) RNAs depicted in (E) were analyzed for generation of eRL by RT-PCR. Left panel combines three independent experiments each in technical duplicates (six measurements per data point ± S.D), *** P < 0.001. Right panel depicts a representative fragment analysis in a 12% PAA gel stained with SYBRGold. One individual experiment of three independent experiments is shown. In vitro RNase A-digested HEK293 RNA served as positive control.
