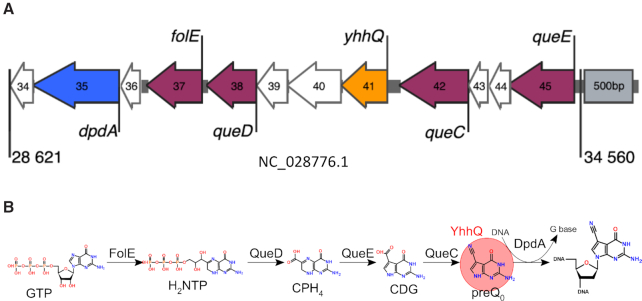
Figure 1.
(A) The 7-deazaguanine modification gene cluster in CAjan bacteriophage (NC_028776.1) located in the region 28 621–34 560 bp. Genes are colored by function: purple are genes involved in preQ0 synthesis, orange is the gene coding for the preQ0 transporter, blue is the dpdA gene and in white are genes coding for unknown functions. Numbers correspond to gene product numbers from NC_028776. On the right side, there is a 500 bp grey rectangle for size reference and rest of the Figure is drawn to this scale. (B) Biosynthesis pathway of preQ0. The biosynthesis starts with GTP which is transformed to preQ0 by enzymes FolE, QueD, QueE and QueC. Alternatively, YhhQ can transport preQ0 from the outside. The last step is DpdA enzyme that exchanges targeted G bases with the preQ0.