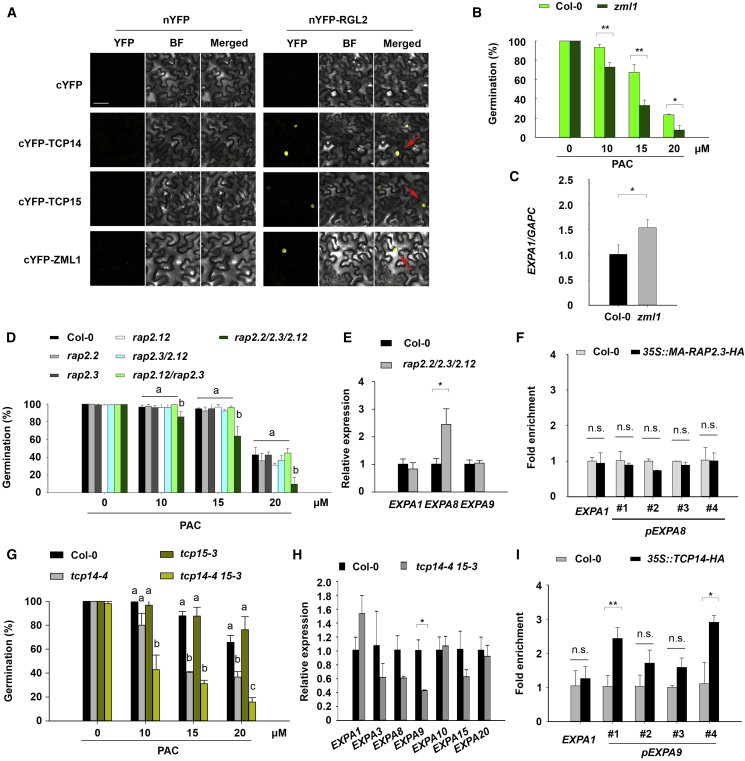
Figure 5.
Functional Validation of the Signal Integration Network
(A) BiFC assay investigating the interaction between DELLA and TFs in plant cells. Fusion proteins were co-expressed in tobacco leaves by using Agrobacterium infiltration and images represent co-transfected cells with visible fluorescence. YFP, fluorescence of yellow fluorescent protein; BF, bright field; merged, merger of the YFP and bright-field images. Red arrows indicate the position of YFP signal. The scale bar indicates 50 μm.
(B) Germination of zml1 imbibed in different concentrations of PAC.
(C) Relative expression of the ZML1 target EXPA1 in the zml1 background determined by using qPCR.
(D) Same as (B) using combinations of rap2.2 rap2.3 and rap2.12 mutant seeds.
(E) Relative expression of the ERFVII TF targets EXPA1, EXPA8, and EXPA9 in the rap2.2 rap2.3 rap2.12 mutant background determined by qPCR.
(F) ChIP using HA-tagged RAP2.3 on the EXPA8 promoter. The EXPA1-coding region was used as a negative control and no specific signals were observed in the negative-control region.
(G) Same as (B) using tcp14 and tcp15 mutant seeds.
(H) qPCR analysis of EXPA genes targeted by TCP14 and TCP15.
(I) ChIP using HA-tagged TCP14 on the EXPA9 promoter. The EXPA1-coding region is included as a control.
Data in (B), (C), (E), (F), (H), and (I) were statistically analyzed by using Student’s t test (∗p < 0.05, ∗∗p < 0.01). Statistically significant differences in (D) and (G) are denoted with different lowercase letters (one-way ANOVA with Tukey post hoc test, p < 0.05). Error bars represent SD from three independent biological repeats.
See also Figure S5.