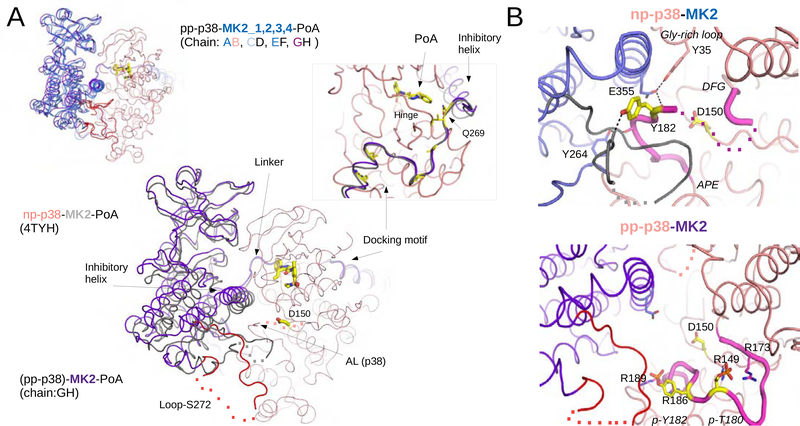
Figure 6. Crystal structure of the pp-p38-MK2-PoA ternary complex.
(A) Comparison of the nonphosphorylated p38-MK2-PoA (PDB ID: 4TYH) and the pp-p38-MK2-PoA crystal structures. The four heterodimers from the pp-p38 complex were superimposed through the MAPK (colored in salmon) and MK2 molecules are colored in blue (panel on the left corner). One of the heterodimers from this complex was similarly superimposed to the nonphosphorylated heterodimer (main panel). The PoA compound and the p38 catalytic site (Asp150) is colored in yellow, and the flexible loop region containing the Ser272 regulatory site in MK2 is colored red. Dotted lines indicate disordered regions. The inset on the right shows a zoomed-in view of the Docking motif-Linker-Inhibitory helix region of MK2, but the view is 180º vertically rotated compared to the other panels. Key contact residues, responsible for high affinity p38-MK2 binding are colored yellow (e.g. Ile730, Ile732, Ile375, Leu382 and Arg386). In addition, a pivotal residue in the Linker connecting the docking motif and the inhibitory helix is also highlighted (Gln269). This region serves as a pivot through which the kinase domains may bind differently but still held together similarly by the D-motif mediated interaction. Notice that the p38 Hinge (connecting the N- and C-lobes of the MAPK) and the MK2 Linker both contribute to PoA inhibitor binding.
(B) Structural comparison of the nonphosphorylated vs phosphorylated p38 activation loop. Panels on the top or below show a zoomed-in view around the p38 catalytic site (Asp150) from the nonphosphorylated p38-MK2-PoA ternary complex (PDB ID: 4TYH) or from the activated pp-p38-MK2-PoA complex, respectively. Notice that the nonphosphorylated p38 activation loop is wedged in-between the MK2 inhibitory helix and the structured part of Loop-S272. The activation loop is fixed in this inhibitory position by several H-bonds (shown with dashed lines). The activation loop of p38 between the DFG loop and the APE motif are colored in magenta. Disordered segments are indicated with a dotted line. Phosphates of the double-phosphorylated p38 AL (p-Thr180 and p-Tyr182) are coordinated by arginines (Arg149, Arg173, Arg186 and Arg189), which pulls the AL down and opens up the catalytic site. See also Figure S7.