Figure 1.
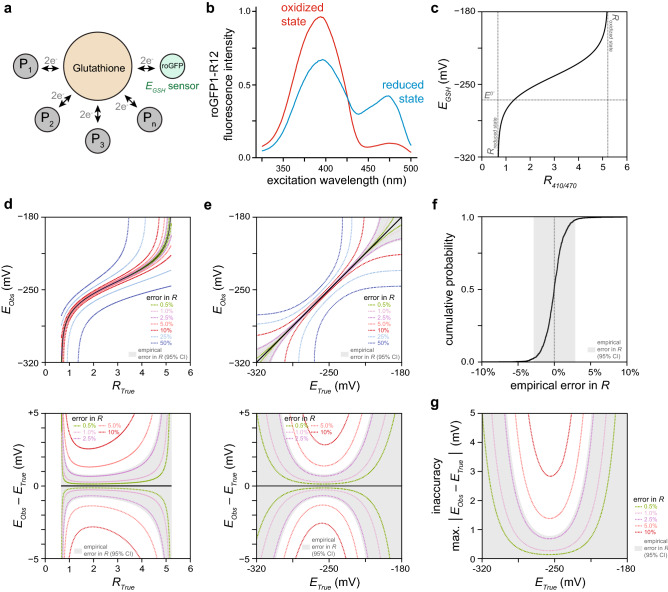
Determining the range of glutathione redox potential EGSH values we can measure accurately with the roGFP1-R12 biosensor. (a) Glutathione redox potential (EGSH) directs the oxidation of cysteines in hundreds of proteins in the same direction, resulting in their concerted regulation. (b) The reduced and oxidized states of the roGFP1-R12 biosensor have different fluorescence spectra8, enabling EGSH measurement via R (fluorescence ratio) microscopy. (c) The conversion map from R to EGSH is highly nonlinear. Rreduced state and Roxidized state refer to the ratiometric emission of ensembles of reduced and oxidized biosensors, respectively. E0′ is the standard midpoint potential of the biosensor. (d) The top panel shows how measurement errors in R cause observed EGSH values (EObs) to differ from the true EGSH values (ETrue) that would be observed if R was measured with no error (RTrue). The bottom panel shows how the size of an EGSH error (EObs – ETrue) depends not only on the size of the error in R but also on the value of R. Each dotted curve corresponds to a different fold-change error in R. The shaded region corresponds the interval encompassing 95% of the predicted EObs values for each RTrue value, given our empirical error in R. (e) Transforming the map from RTrue to ETrue in the top and bottom panels shown in (d) produces plots showing how errors in R influence the map from ETrue to EObs (top panel) and how the size of an EGSH error depends not only on the size of the error in R but also on the value of ETrue (bottom panel). Each dotted curve corresponds to a different fold-change error in R. The shaded region shows the interval encompassing 95% of the predicted EObs values for each ETrue value, given our empirical error in R. (f) Cumulative distribution of the empirical fold error in R in live C. elegans expressing the roGFP1-R12 biosensor in the cytosol of the anterior (pm3) muscles of the pharynx, the feeding organ. This error distribution was obtained by aggregating with equal weight the empirical fold error in R of five separate experiments (see Supplementary Note 3). 95% of the errors in R fall within the interval (− 2.8%, + 2.8%), shown shaded in gray. This interval quantifies the precision of our fluorescence-ratio measurements. (g) EGSH measurement inaccuracy (the maximum absolute difference between ETrue and EObs) decreases with increased precision of R measurement. Each dotted curve corresponds to a different precision of R measurement. The shaded region shows the interval encompassing 95% of the predicted EGSH measurement inaccuracies for each ETrue value, given our empirical error in R.