Abstract
COVID-19 an outbreak of a novel corona virus originating from Wuhan, China in December 2019 has now spread across the entire world and has been declared a pandemic by WHO. Angiotensin converting enzyme 2 (ACE2) is a receptor protein that interacts with the spike glycoprotein of the host to facilitate the entry of coronavirus (SARS-CoV-2) hence causing the disease (COVID-19). Our experimental design is based on bioinformatics approach that combines sequence, structure and consensus based tools to label a protein coding single nucleotide polymorphism (SNP) as damaging/deleterious or neutral. The interaction of wildtype ACE2-spike glycoprotein and their variants were analyzed using docking studies. The mutations W461R, G405E and F588S in ACE2 receptor protein and population specific mutations P391S, C12S and G1223A in the spike glycoprotein were predicted as highly destabilizing to the structure of the bound complex. So far, no extensive in silico study has been reported that identifies the effect of SNPs on Spike glycoprotein-ACE2 interaction exploring both sequence and structural features. To this end, this study conducted an in-depth analysis that facilitates in identifying the mutations that blocks the interaction of two proteins that can result in stopping the virus from entering the host cell.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, 2019-nCOV, ACE2, spike glycoprotein, SNPs
Introduction
COVID-19 an outbreak of a novel corona virus originating from Wuhan, China in December 2019 has now spread across the entire world and has been declared a pandemic by WHO. The SARS-CoV-2 belongs to the family of corona viruses that caused severe acute respiratory syndrome (SARS) in the year 2002 and can be transmitted from animals to humans. The human-human transmission is currently prevailing, increasing the number of total patients to 7,127,753 by June 10th, 2020 including 409,150 deaths as reported by WHO (https://www.who.int/). Currently, the only treatment of this disease is self-isolation and several academic and commercial groups are in search of a drug and/or a vaccine for this disease.
The interaction of surface spike glycoproteins of SARS-CoV with the receptor protein Angiotensin converting enzyme II (ACE2) (EC number: EC 3.4.17.23) facilitates the virus entry into the host cell. Spike glycoprotein is a homotrimer and each monomer possess around 1200 amino acids. The receptor binding domain (RBD) is a small domain of spike glycoprotein spanning from 360-575 and from which the residues 424-494 make up the receptor binding motifs which directly activates the interaction with ACE2. Moreover, the residues of spike glycoprotein that interacts with ACE2 are evolutionary conserved residues (Basit et al., 2020; Kalathiya et al., 2020; Yan et al., 2020).
ACE2 is an enzyme that is found on the outer membranes of lung cells, arteries, heart, kidney and intestines and belongs to the renin-angiotensin-aldosterone system (RAAS), which regulates blood pressure and body fluid, hence it has an important contributing role in hypertension and cardiovascular/renal diseases. The protease Renin, cleaves angiotensinogen to create Angiotensin (Ang) I. ACE2 then cleaves Ang I to produce Ang II. Decreasing the level of ACE2 can help fight the infection as this might affect the interaction of two proteins hence stopping the virus entry into the cell (Imai et al., 2008).
Lately, much work has been done in determining the exact cause of COVID-19 disease and in finding its cure by designing vaccines/drugs against it. Haider et al. used pharmacophore based virtual screening for in silico drug design (Haider et al., 2020). The authors performed molecular docking analysis and identified three potential drugs ZINC20291569, ZINC90403206, and ZINC95480156 that showed the strongest binding interactions against the active site of main protease of SARS-CoV-2. The receptor binding domain (RBD) of spike glycoprotein has been reported to interact with ACE2 receptor protein (Othman et al., 2020). This study also identified the importance of Q493, P499 residues in the RBD which are important for maintaining the stability of bound complex (Othman et al., 2020). The study carried out by Peele et al. designed an in-silico based multi-epitope vaccine that activates both CD4 and CD8 immune response cells. The B and T cell epitopes were used to design this vaccine that are responsible for provoking immune response in the host cell. Further, the authors have performed molecular docking and simulation studies which confirmed the protein-protein interactions and stability of the binding pose (Ap & Vs, 2020). Recently, few studies have conducted computational analysis on medicinal plant to design a treatment against coronavirus. The study carried out by Sinha et al. used Saikosaponins as a treatment for COVID-19. It belongs to a group of oleanane derivates and possess various antiviral and anti-inflammatory activities. The molecular docking studies were carried out on nsp15 (PDB ID: 6W01) and 2019-nCoV spike glycoprotein (PDB ID: 6VSB). The resulting binding affinities showed the Saikosaponins binds really well with both of the proteins and can be served as a new future molecule for treating COVID-19 (Sinha et al., 2020).
The complex of ACE2 with B0AT1 that stabilizes the ACE2 structure was studied with spike glycoprotein. The authors (Armijos-Jaramillo et al., 2020) have identified the binding affinities of the RBD of ACE2 with the extracellular peptidase domain (PD) of spike glycoprotein and how the mutations affect the bound complex. The structural interpretation may enlighten the mechanisms of viral infection and helps in developing antiviral therapeutics. Another study explored the evolutionary dynamics of COVID-19 disease by exploiting the spike glycoprotein interaction with ACE2 (Yan et al., 2020). The evolutionary analysis was conducted for spike glycoprotein to determine if the genomic regions are under purifying selection. The authors discovered few residues inside the RBD that are highly conserved and are also thought to provide stabilizing interactions in the bound complex (nCoV-ACE2) (Ji et al., 2020). A recent study has conducted a phylogenetic based evolutionary analysis to determine the common ancestor by carrying a comparative analysis on genomic sequence similarity from humans, bat, snakes and mice. The analysis predicted that snake is the major reservoir of this disease also, there is a homologous recombination present in the RBD of spike glycoprotein which may justify the transmission of the disease from animals to humans (The UniProt Consortium, 2017). Also, an experimental study conducted by Wooster et al (Wooster et al., 2020) analyzed the variants in ACE2 in 62 COVID-19 positive patients. The genotyping was performed using the Illumina Infinium MEGA Consortium v1 SNP array. GTEx was used to determine significant SNP associated with ACE2. Out of 10 eQTL variants, 6 were significantly associated with hospitalization requirements for COVID-19. This study provides a genetic link between ACE2 genotype and its severity with the disease formation. Another, population specific study carried out by Khayat et al (Khayat et al., 2020) used exomics analysis to study the polymorphism in native and mixed American populations. The data was gathered from 1000 Genomes Phase 3 database which contains the data from 26 different populations. They have identified three polymorphisms rs147311723 (L731F), rs142017934 and rs4646140 that are common in African population. Among these the rs142017934 variant was observed as more damaging and can affect the translation of ACE2 gene, increasing the expression of this gene. Moreover, higher frequencies of rs1027571965 (A673G), rs889263894 (K541I), rs2285666 and rs35803318 were observed in the American population. These SNPs increased the ACE2 expression level in the brain tissues. This particular population group has genetic distinctiveness and was also less exposed to the viral infections. Calcagnile et al. also found polymorphisms in ACE2 gene in African and European populations by carrying out docking and simulations based study. The study reported two variants S19P in African and K26R in European population as significantly associated with ACE2 expression and it’s binding with spike glycoprotein. S19P is predicted to decrease the binding affinity, while K26R is predicted to increase the binding affinity of ACE2 with spike glycoprotein (Calcagnile et al., 2020).
Understanding the fundamentals of virus entry into the host cell including the protein interactions that facilitates this process is the key step that can lead to the development of vaccines and drugs. We have employed a computational approach to determine the nsSNPs in ACE2 and to determine which variants are more crucial in disrupting the structure-function of the ACE2 protein and its interaction with the 2019-nCoV spike glycoprotein. So far, no extensive study has been reported that identifies the effects of SNPs on 2019-nCoV-ACE2 interaction by exploring both the sequence and structural features. To this end, this study conducted an in-depth analysis that facilitates the identification of the mutations that are affecting the interactions of two proteins, and might be responsible for preventing infections. Our study will contribute effectively in providing better understanding of disease mechanisms.
Materials and methods
Sequence retrieval and data mining of SNPs
The protein sequences and structures of nCoV spike glycoprotein (PDB ID: 6VSB) and Angiotensin converting enzyme 2 ACE2 (PDB ID: 1R42) were downloaded from UniProt (https://www.uniprot.org/) (Apweiler et al., 2004) and Protein Data Bank (Kouranov et al., 2006). Further, the genomAD database was queried to obtain the human ACE2 missense SNPs (https://gnomad.broadinstitute.org/) (Karczewski et al., 2020). After redundancy reduction a total of 230 SNPs were retrieved for further analysis.
Functional annotation
The downloaded SNPs were further subjected to functional annotation analysis using several bioinformatics tools. The first group of tools includes sequence homology based tools: SNPNexus (SIFT- PolyPhen) (Dayem Ullah et al., 2012), Protein Variation Effect Analyzer (PROVEAN) (Choi & Chan, 2015) and Mutation Accessor (Reva et al., 2011). The second group includes consensus based methods: Meta-SNP (Bendl et al., 2014), SNPs&GO (Capriotti et al., 2013) and PredictSNP (Bendl et al., 2014). The SNPNexus has built-in SIFT and PolyPhen tools, as these are sequence homology based tools they first obtain the homologous sequences of the query sequence and perform the multiple sequence alignment. The score of 0-0.5 classifies the mutation as damaging or deleterious. PROVEAN webserver takes in protein sequence along with the list of mutations to run a homology search via BLAST and constructs multiple sequence alignment. The PROVEAN scores have a cutoff threshold of −2.5 for labelling the mutation as neutral and deleterious otherwise. The multiple sequence alignment built by Mutation Accessor determines the evolutionary conservation profile of the query protein. Mutation Assessor uses combinatorial entropy optimization to determine the functionally important residues by clustering them into subfamilies. The residues were classified into specificity residues, conserved residues or neutral residues.
The Meta-SNP, PredictSNP and SNPS&GO are consensus based methods that incorporate various tools. Meta-SNP is a random forest based binary classifier that combines four predictors SNAP (Screening for Non-Acceptable Polymorphism) (Bromberg & Rost, 2007), SIFT (Sorting Intolerant from Tolerant), PANTHER (Protein Analysis through Evolutionary Relationships) (Thomas & Kejariwal, 2004) and PHD-SNP (Predictor of Human Deleterious SNP) (Capriotti & Fariselli, 2017). PHD-SNP identifies if the particular SNP is disease associated or neutral while the SNAP, PANTHER and SIFT functionally annotate the variants. PredictSNP has eight tools nsSNPAnalyzer (Bao et al., 2005), PolyPhen (Polymorphism Phenotyping), SNAP, MAPP (Multivariate Analysis of Protein Polymorphism), PHD-SNP, SIFT, and consensus PredictSNP. SNPS&GO implements the PHD-SNP method which is SVM classifier that combines features from sequence, evolutionary information and functional features derived from GO terms. A SNP is labelled as high risk if it is predicted as deleterious from 5 out of these 7 tools for obtaining high confidence results.
Predicting evolutionary conserved residues
The ConSurf webserver (Celniker et al., 2013) was used to predict the evolutionary conserved residues and also to identify conserved motif patterns (if any). The FASTA protein sequence of ACE2 was sent as input to ConSurf, the tool then generates the multiple sequence alignment automatically and also builds the 3D model of the protein. Bayesian algorithm was used to compute the conservation score by performing phylogenetics analysis between the homologous sequences. The scores in the range of 1-4 are annotated as variable, 5-6 as intermediate and 7-9 as conserved. Furthermore, the tool also predicts if the particular residue is buried or exposed which can further reveal the structural and functional importance of that residue. The tool is available at http://consurf.tau.ac.il/2016/.
In order to determine any population specific mutation we have downloaded the genomic sequences of the new coronavirus of different origins from the Global Initiative on Sharing Avian Influenza Data (GISAID) database (Shu & McCauley, 2017). Further detail is provided in Supplementary File S1. The sequences were sent to MAFFT for multiple sequence alignment (Katoh et al., 2005).
Analyzing protein stability
We have used mCSM webserver to predict the protein stability analysis of the observed mutants (Pires et al., 2014). The server is based on graph based signatures which are calculated as distance patterns around the wild type residue. Given a mutation site the algorithm first creates the wild type environment by defining a distance of atoms from its geometric mean. A matrix is created which defines the pairwise distance between the two atoms. Furthermore, the pharmacophore count was also added to the feature set. The difference of Gibbs free energy DDG between wildtype and mutant residue was analyzed which then labels a mutation as destabilizer or neutral.
Predicting disease related mutations using MutPred
To predict the disease associated SNPs we have used MutPred (Mort et al., 2014) that classifies a variant as pathogenic (disease associated) or neutral by using structural, functional and evolutionary features. It also includes tools to predict structural disorder (TMHMM, MARCOIL, and DisProt) that determines the molecular basis of the pathogenicity associated with the amino acid substitutions. A consensus based approach is likely to bring a high confidence prediction score.
Protein 3D structure modelling
The deposited 3D structures of ACE2 and 2019-nCoV spike glycoprotein were downloaded from RCSB PDB. We used I-TASSER (Zhang, 2008) to generate complete 3D structure of both proteins for their wild-type and mutated sequences, as the resolved structures do not cover the full amino acid sequence of the proteins. I-TASSER is an automated homology modeling based tool that combines threading and ab initio structure prediction. The quality of the generated protein model was analyzed by ERRAT (Colovos & Yeates, 1993). The wild-type and mutant models were superimposed and visualized in Chimera 1.11 (Pettersen et al., 2004).The structural similarity of the models was then analyzed using TM-Align (Zhang & Skolnick, 2005).
Molecular docking analysis
To predict the protein complex between the two interacting proteins, we have used ClusPro webserver (Kozakov et al., 2017) by using the default settings. The tool is based on a Fast Fourier Transform Correlation approach that makes it very flexible for computing billions of docked molecules by using a simple scoring function. ClusPro rotated the ligand in 70,000 different orientations and 1000 poses were then picked up based on the lowest scores. The algorithm behind ClusPro further used greedy approach to generate clusters with a 9 Å C-alpha RMSD radius. The server outputs 10 different docked poses which were ranked according to the energy and cluster size. The 2019-nCoV spike glycoprotein was docked with its receptor protein ACE2. Both the wild type ACE2 and the mutant models of ACE2 were docked with 2019-nCoV spike glycoprotein wildtype and mutant proteins to analyze the effect of variants on the interaction of the two proteins.
Results
Retrieval of non-synonymous (nsSNPs) in ACE2
Total of 230 missense SNPs in ACE2 were downloaded from genomAD database. From these missense SNPs, SIFT and PolyPhen predicted 145 SNPs as damaging/deleterious which were further taken to PROVEAN server. From these 145 SNPs, PROVEAN identified 92 mutations as deleterious/damaging. The functional effect of the nsSNPs was further analyzed by the Mutation Accessor, Meta-SNP, SNPs&GO, and Predict-SNP. To predict with high confidence, we have labelled a SNP as damaging only if it is predicted as deleterious/damaging from at least 5 out of 7 tools. Based on this, 34 candidate SNPs were selected as deleterious which were predicted commonly as damaging from the 7 tools mentioned above. Detailed predictions of the SNPs for each tool are tabulated in Supplementary File Table S2, S3 S4 and S5. Figure 1 represents the general workflow of the methodology and (Table 1) summarizes the results obtained from functional annotation tools..
Figure 1.
An overview of the methodology design of nsSNPs structure-function analysis.
Table 1.
Combined deleterious SNPs predicted from SNPNexus, PROVEAN, Meta-SNP, PredictSNP, SNPs&Go and Mutation Accessor.
| Mutation | SNPNexus | PROVEAN | Meta-SNP | PredictSNP | SNPs&GO | Mutation Accessor | Result |
|---|---|---|---|---|---|---|---|
| L595V | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| F588S | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| L585P | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| L570S | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| S563L | Deleterious | Deleterious | Neutral | Deleterious | Deleterious | Deleterious | Deleterious |
| S547F | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| S547C | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| I544N | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| K541I | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| V506A | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| F504L | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| F504I | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| G466W | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| W461R | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| G405E | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| N397D | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| G377E | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| E375D | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| M360L | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| D355N | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| E312K | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| P263S | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| Y252C | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| A242V | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| Y207C | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| R204T | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| A191P | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| L186S | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| V184G | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| V184A | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| P178L | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| C141Y | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
| S128T | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious |
Conservation profile of SNPs
The 34 candidate SNPs predicted as damaging/deleterious from the functional annotation tools are further subjected to evolutionary conservation analysis by ConSurf. We have considered those SNPs that have scores in the range of 7-9 to pick the highly conserved mutations. From the 34 mutations, 21 are found to be conserved and buried and present at the core of the protein, and 13 are conserved and present at the surface of the protein. The detailed results are provided in Supplementary File Table S6.
Further, the multiple sequence alignment was performed on the genomic sequences of spike glycoprotein sequences sampled from GSAID between December 2019 and April 2020. We found mutations P391S, R207C, and P2965L occurring in the Pakistani population, variant G1223A in UK specific population and C12S in Indian population. All these variants are occurring at evolutionary conserved regions as predicted by ConSurf with R207C predicted to be buried hence contributing structurally while the variants P391S, P2965L, G1223A and C12S are predicted as functionally important residues.
Predicting protein stability
The change in protein stability was predicted from mCSM webserver. The change in Gibbs free energy determines if the mutation at particular residue is disrupting the stability of the protein. The results from mCSM showed that a few mutants are highly destabilizing to the structure of the protein hence expected to disrupt the proper functioning of the protein. These variants were found to be evolutionary conserved using ConSurf. The results of the significant mutants are tabulated in Table 2 while detailed results are presented in Supplementary Table S7.
Table 2.
Damaging variants predicted from mCSM and Consurf.
| Mutation | DDG | Effect | Evolutionary Conservation Analysis |
|---|---|---|---|
| F588S | −3.33 | Highly Destabilizing | Buried – Highly Conserved |
| V573A | −2.14 | Highly Destabilizing | Buried- Less conserved |
| L570S | −3.03 | Highly Destabilizing | Buried- Highly conserved |
| Y521H | −2.41 | Highly Destabilizing | Buried- Less conserved |
| V506A | −2.40 | Highly Destabilizing | Buried- Less conserved |
| W461R | −2.14 | Highly Destabilizing | Buried- Highly conserved |
| G405E | −2.94 | Highly Destabilizing | Buried- Highly conserved |
| G377E | −2.23 | Highly Destabilizing | Buried- Highly conserved |
| P263S | −3.44 | Highly Destabilizing | Exposed- Highly conserved |
| R204T | −2.06 | Highly Destabilizing | Exposed- Conserved |
| L186S | −2.99 | Highly Destabilizing | Buried- Conserved |
| V184G | −2.19 | Highly Destabilizing | Buried- Conserved |
| A164S | −2.16 | Highly Destabilizing | Buried- Conserved |
| R207C | −1.42 | Destabilizing | Buried- Highly conserved |
| P391S | −1.22 | Destabilizing | Exposed- Highly conserved |
| P2965L | −1.01 | Destabilizing | Exposed- Highly conserved |
| G1223A | −1.42 | Destabilizing | Exposed- highly conserved |
| C12S | −1.92 | Destabilizing | Exposed- highly conserved |
Predicting disease associated SNPs using MutPred
The SNPs predicted as potential destabilizers were further checked with MutPred to predict their association with disease. MutPred predicts the change in the molecular processes upon mutation like alterations of transmembrane helices, altered disordered interface, gain or loss of catalytic sites, solvent accessibility and post translational modifications. The scores that have a p-value < 0.05 and g-value > 0.75 are considered as high risk SNPs. The results of MutPred showed that the F588S, A191P, I544N, V184G, L186S, G405E and L585P substitutions had the highest g-value. These mutants were also predicted as deleterious from the functional annotation tools. Moreover, the mutants F588S, A191P, I544N, V184G, and G405E were also predicted as evolutionary conserved residues. The results are tabulated in Supplementary Table S8.
3D Structure modelling of ACE2 and molecular docking analysis
The experimentally resolved 3D structure of ACE2 (PDB ID: 1R42) contains residues from 1-615, however the full protein length of ACE2 is 805 amino acids. Therefore, the complete tertiary structure was generated using I-TASSER. The mutant models of high risk SNPs were also generated using I-TASSER. Furthermore, the structural similarity between the wild-type protein and its variants were calculated using TM-Align. Three mutants (F588S, G405E and W461R) showed the highest deviation with RMSD 1.86, 1.45 and 1.46 Å, respectively. The rest of mutants either showed very less deviation or no deviation.
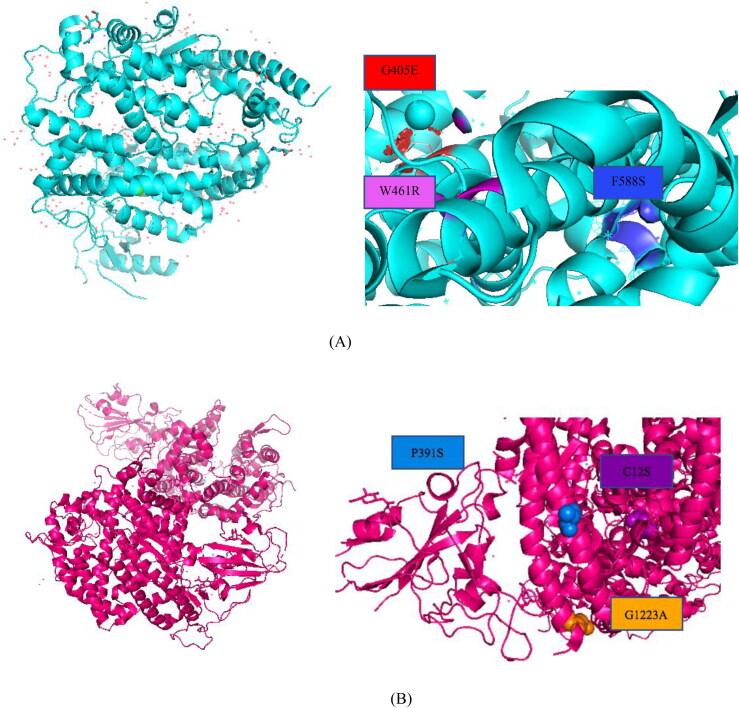
Further, from the multiple sequence alignment we have identified few variants among Pakistani (P391S), Indian (C12S) and England (G1223A) populations. The mutant models of P391S, C12S, and G1223A showed significant deviation with RMSDs of 1.56, 1.35 and 1.46 Å. The mutant models were superimposed with the wild type protein model using Chimera to identify the mutation positions. The quality of the protein structures were checked with ERRAT which showed the value of 80.83 for the wild type ACE2, 95.6 for F588S, 90.53 for G405E and 95.06 for W461R mutant. Similarly, we obtained 87.45 for wildtype spike glycoprotein, 91.36 for P391S, 97.34 for C12S and for 97.26 for G1223A mutant. The results are figured in Figure 2.
Figure 2.
(A) Structural position of the ACE2 mutants G405E, F588S and W461R in the protein 3D structure visualized in PyMol. (B) Structural position of population specific mutations of spike glycoprotein, P391S, C12S and G1223A in the protein 3D model visualized in PyMol.
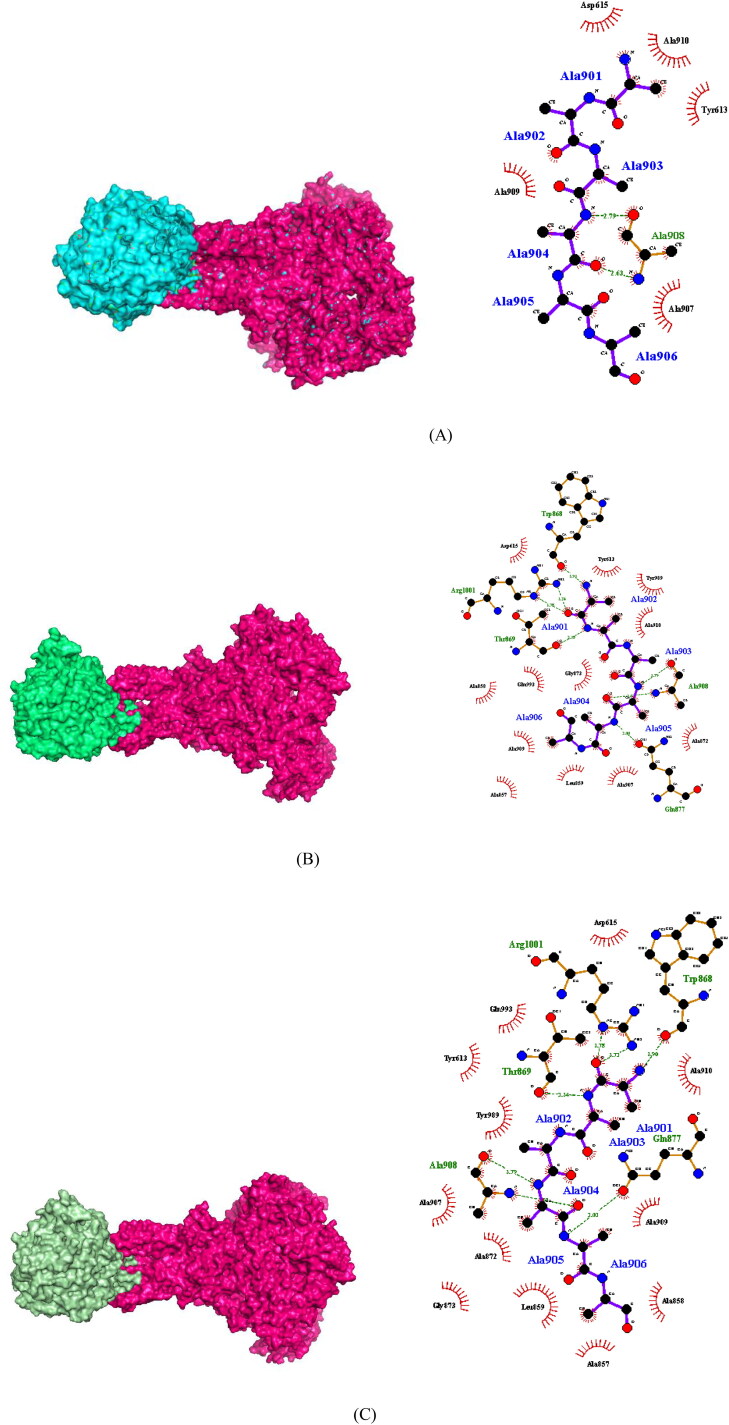
In order to determine how strongly the proteins are bound together in their wild form and how the mutations are affecting this bound structure, we used the ClusPro protein docking webserver. We selected cluster 1 because of its large size and lowest energy. Furthermore, we calculated the buried surface area (BSA) using PyMol to interpret the protein-protein interactions (1R42-6VSB). We considered buried surface area (BSA) as the measure of strength of two interacting proteins. The bound complex of wildtype spike glycoprotein with ACE2 is expected to be more stable with the highest BSA. The results obtained showed that the mutations are noticeably disrupting the bound complex. The Table 3 and Figure 3 shows the buried surface area of the docked complexes of wild type and mutant models. Also, the docked structures were analyzed via LIGPLOT+ (Laskowski and Swindells, 2011) to check the interacting binding residues of nCoV-ACE2. The binding pocket of wild type docked complexes and the mutant models showed different set of residues with mutant F588S showed complete loss of binding pocket hence, is largely effecting the interaction between the two proteins as figured in Figure 3.
Table 3.
Predicted buried surface area of the two interacting proteins with wildtype ACE2 and with the mutant models of ACE2.
| Interacting Complex | ClusPro Buried SA (A2) |
|---|---|
| 6VSB-1R42 (Wildtype) | 3558 |
| 6VSB-1R42 (G405E) | 2952 |
| 6VSB-1R42 (W461R) | 2958 |
| 6VSB-1R42 (F588S) | 2854 |
| 6VSB (P391S)- 1R42 | 2980 |
| 6VSB (C12S)- 1R42 | 2760 |
| 6VSB (G1223A)- 1R42 | 3215 |
Figure 3.
(A) Docked complex of wild type 2019-nCoV spike glycoprotein with wild type ACE2 receptor protein. (B) The docked complex of 2019-nCoV spike glycoprotein with mutant G405E ACE2 receptor. (C) The docked complex of 2019-nCoV spike glycoprotein with mutant W461R ACE2 receptor.(D) The docked complex of 2019-nCoV spike glycoprotein with mutant F588S ACE2 receptor. (E) The docked complex of mutant 2019-nCoV spike glycoprotein P391S with wild type ACE2. (F) The docked complex of mutant 2019-nCoV spike glycoprotein C12S with wild type ACE2. (G) The docked complex of mutant 2019-nCoV spike glycoprotein G1223A with wild type ACE2.
In addition to that, the molecular docking studies were validated on electron microscopic crystal structures of spike glycoprotein and ACE2 proteins (PDB IDs: 6ACJ, 6ACG, 6CS2, 6ACK). We separated the ligand from the receptor in the electron microscope structure and then docked the ligand on to the receptor using our above mentioned protocol. We were able to capture the binding pose as demonstrated by the similarity of the buried surface area values. The buried surface areas computed on the structures 6CS2, 6ACJ and 6ACK showed the similar area to our modeled structures. These were also analyzed with the mutant models of receptor-ligand (PDB: 1R42, 6VSB) and the decrease in BSA was observed in the mutants as compared to the wildtype docked structures.
The PDB structures 6ACG and 6ACJ are Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein and ACE2 complex, ACE2-bound conformation 1 and ACE2-bound conformation 2. The two conformations of ACE2 showed different BSA, the ligand docked with conformation 2 demonstrated similar trends in BSA as of our docked structures (1R42-6VSB). The results are tabulated in Table 4.
Table 4.
The buried surface area of the wild and mutant models in experimental PDB structures.
| Interacting Complex | Wild-Type Buried SA (A2) | Wildtype Re- docked. Buried SA (A2) 1R42 | 1R42- F588S Buried SA (A2) | 1R42- W461R Buried SA (A2) | 1R42- G405E Buried SA (A2) | Wildtype Re- docked. Buried SA (A2) 6VSB | 6VSB -P391S Buried SA (A2) | 6VSB -C12S Buried SA (A2) | 6VSB -G1223A Buried SA (A2) |
|---|---|---|---|---|---|---|---|---|---|
| 6ACJ | 3549 | 3550 | 2756 | 2759 | 3559 | 2980 | 2788 | 2485 | 2958 |
| 6CS2 | 3562 | 3560 | 2547 | 2872 | 2972 | 3534 | 2218 | 3077 | 3402 |
| 6ACG | 3045 | 3717 | 3722 | 3723 | 3723 | 4473 | 4435 | 4546 | 4479 |
| 6ACK | 3552 | 3569 | 3438 | 3426 | 3443 | 3542 | 2123 | 3290 | 3421 |
The mutant residue in G405E is neutral, bigger in size and is more hydrophobic than the wild type residue. The mutation is occurring at the conserved region and is also a part of domain Peptidase M2, Peptidyl-Dipeptidase A which is important for the main activity of the protein and mutation can lead to function disorder. There is a difference of charge as mutated residue is negatively charged this can cause repulsion of ligands or other residues with the same charge. The hydrophobicity of the wild-type and mutant residue differs therefore, the hydrophobic interactions either in the core of the protein or on the surface will be lost. The mutation W461R also introduces the difference in charge between the wild-type and mutant amino acid, which can cause repulsion of ligands or other residues with the same charge. The wild-type and mutant amino acids differs in size. The mutant residue is smaller, this might leads to loss of interactions.
For the mutation F588S the hydrophobicity of the wild-type and mutant residue differs. Hydrophobic interactions, either in the core of the protein or on the surface, will be lost. Only this residue type was found at this position in multiple sequence alignment as predicted from HOPE webserver. Mutation occurring at a 100% conserved residue is usually damaging for the protein. Based on this conservation information this mutation is probably damaging to the protein (Venselaar et al., 2010).
The multiple sequence alignment of the genomic sequences predicted few population specific mutations. The docked complexes of P391S from Pakistani population, C12S from Indian population and G1223A from England population showed the difference of interaction between the 2019-nCoV spike glycoprotein with its receptor ACE2 protein. These mutations expected to disrupt the bound protein complex hence blocking the interaction between the two proteins.
Discussion
Single nucleotide polymorphisms (SNPs) refers to a change in a single residue at a particular position in a sequence which occurs at a frequency of more than 1% in a population. SNPs can occur at both coding and non-coding regions of the genome among which, coding SNPs have a high potential of having a drastic effect on protein structure and function (Khalid & Sezerman, 2020). A large amount of the data has been deposited in the databases on non-synonymous SNPs (nsSNPs) which has been in the limelight of the current research. Identifying the nsSNPs has various implications in the medical research as these SNPs are associated with diseases which can facilitate drug discovery.
Lately, many studies have been reported on nsSNPs to predict structurally and functionally important variations that are associated with disease formation. The features included in these studies are either sequence related or structure related or they can belong to both categories. This study focused on carrying out a comprehensive study on the SNPs associated with nCoV-ACE2 interactions. We have combined various computational tools to validate the predictions. First, all the nsSNPs were functionally annotated and were then passed to evolutionary conservation analysis. The shortlisted SNPs were then checked for their effect on protein stability. The SNPs that are highly destabilizing to the structure of the protein were further subjected to docking studies. The docking studies were carried out with ClusPro for both the wildtype protein and with the mutant models of ACE2. To check the strength of interaction of the bound complex we have computed buried surface area of the protein complex. We have calculated the values for both wild type and mutant models and have taken this as a measure of strength of interactions between the two proteins. The computed values showed a difference of BSA between the wildtype interaction and interaction of spike glycoprotein with the mutant models of ACE2. A larger variation has observed with the F588S mutant model of ACE2. All these variants were also predicted as highly conserved by the CONSURF webserver hence are expected to be destabilizing for the structure of the protein. This shows that particularly this mutation is affecting the interaction of the two proteins. The spike protein interacts with its receptor protein ACE2 which then facilitates the entry of the virus in the host cell. As, the mutations are dominantly affecting the interactions this further may stop the entry of the virus in the host cell.
In addition to that, mutations predicted from multiple sequence alignment (MSA) of spike glycoprotein were also analyzed. MSA identified P391S, R207C, P2965L in Pakistani population, C12S in Indian population while N1223R in English population. These mutants were checked with the functional annotation tools and all of them predicted these mutations as either benign or neutral but the variants are evolutionary conserved as predicted from ConSurf. Also, when analyzed with mCSM webserver to check if the mutants are affecting the protein stability the scores showed that all mutants (P391S, R207C, P2965L, C12S and G1223A) are destabilizing the structure and function of the protein. Further, to validate this we have utilized HOPE webserver and it was observed for R207C the size difference and the difference of hydrophobicity between the two residues will affect the hydrogen bond formation because the mutant residue might not fit in well to make hydrogen bond. The variant P2965L replaces prolines with leucine, proline is a rigid amino acid that provides rigidity and provides the special conformation of the protein which might be required hence, leucine can disrupt this conformation. Further this residue is exposed at the surface of the protein and have a functional role as predicted by ConSurf involves the interaction with other proteins. On the other hand, the mutation P391S has the size difference and change in hydrophobicity will affect the contacts made by the wildtype residue. Consequently, will affect the hydrophobic interactions. As prolines are special amino acids which are rigid in nature therefore, provide the special conformation which will be disrupted by the mutant residue (Venselaar et al., 2010). Among these, the highest deviated mutant was P391S, we performed the docking analysis of mutant model with the wild type ACE2 protein model to check if the mutation in spike protein is affecting the interaction between the two molecules. The difference of the Buried SA between wildtype docked complexes and with the mutant P391S showed that the mutations are affecting this interaction. The Pakistani population specific mutation in 2019-nCoV spike glycoprotein shall be validated with experimental analysis in future studies to further enlighten its effect on the interaction and to the structure and function of the protein.
In the C12S variant the wildtype residue is more hydrophobic, this difference might disrupt the interactions with other proteins. The mutation G1233A occurring in English population is replacing Glycine the most flexible residue to Alanine. The torsion angles for this residue are very unusual only glycine is flexible enough to make these torsion angles, mutation into another residue will force the local backbone into an incorrect conformation and will disturb the local structure. The size difference of two residues might abolish the actual function of the protein.
Unlike the study carried out by Calcagnile et al (Calcagnile et al., 2020) that predicted the two population specific variants S19P and K26R as significant polymorphisms in African and European population, our functional annotation analysis predicted these mutations as neutral. Also, when analyzed with CONSURF for identifying if the mutations are occurring at evolutionary conserved sites, the server predicted both of them as non-conserved with a score of 3. These mutations are occurring within a specific population hence, the frequency among different populations could be low for this reason, the in-silico analysis predicted them as silent mutations. As, these variants are not identified in the evolutionary conserved regions of the genome therefore, experimental verifications are required to confirm their significance.
Conclusions
We have carried out an in-silico study to determine the deleterious mutations in ACE2 and spike glycoprotein. The functional annotation identified the variants G405E, W461R, F588S in ACE2 while, P391S, C12S and G1223A in spike glycoprotein as lethal and also evolutionary conserved. These were further checked with the docking studies to determine their effect on the protein bound structure. The computed buried surface area for each model showed the significant decrease in the mutant models as compared to the wildtype structures. The docking studied was also validated by computing BSA on already crystallized docked structures of ACE2-nCOV which demonstrated the similar trends of BSA to our docked structures, thus supporting our docking protocol.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ap, K., & Vs, A. (2020, May). Design of multi-epitope vaccine candidate against SARS-CoV-2: A In-Silico study. Journal of Biomolecular Structure & Dynamics, 1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Natale, D. A., O'Donovan, C., Redaschi, N., & Yeh, L.-S L. (2004). UniProt: The universal protein knowledgebase. Nucleic Acids Research, 32(Database issue), D115–9. 10.1093/nar/gkh131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armijos-Jaramillo, V., Yeager, J., Muslin, C., & Perez-Castillo, Y. (2020). SARS-CoV-2, an evolutionary perspective of interaction with human ACE2 reveals undiscovered amino acids necessary for complex stability. bioRxiv. [DOI] [PMC free article] [PubMed]
- Bao, L., Zhou, M., & Cui, Y. (2005). nsSNPAnalyzer: Identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Research, 33(Web Server issue), W480–2. 10.1093/nar/gki372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit, A., Ali, T., & Rehman, S. U. (2020). Truncated human Angiotensin Converting Enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. Journal of Biomolecular Structure and Dynamics, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendl, J., Stourac, J., Salanda, O., Pavelka, A., Wieben, E. D., Zendulka, J., Brezovsky, J., & Damborsky, J. (2014). PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations . PLoS Comput. Biol, 10(1), e1003440. 10.1371/journal.pcbi.1003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg, Y., & Rost, B. (2007). SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res, 35(11), 3823–3835. 10.1093/nar/gkm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagnile, M., Forgez, P., Iannelli, A., Bucci, C., Alifano, M., & Alifano, (2020). bioRxiv.
- Capriotti, E., Calabrese, R., Fariselli, P., Martelli, P. L., Altman, R. B., & Casadio, R. (2013). WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics, 14(Suppl 3), S6. 10.1186/1471-2164-14-S3-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti, E., & Fariselli, P. (2017). PhD-SNPg: A webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Research, 45(W1), W247–52. 10.1093/nar/gkx369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, G., Nimrod, G., Ashkenazy, H., Glaser, F., Martz, E., Mayrose, I., Pupko, T., & Ben, ‐T. N. (2013). ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Israel Journal of Chemistry, 53(3-4), 199–206. 10.1002/ijch.201200096 [DOI] [Google Scholar]
- Choi, Y., & Chan, A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England), 31(16), 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: Patterns of nonbonded atomic interactions . Protein Sci, 2(9), 1511–1519. Sep 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah, A. Z., Lemoine, N. R., & Chelala, C. (2012). SNPnexus: A web server for functional annotation of novel and publicly known genetic variants (2012 update). Nucleic Acids Research, 40(Web Server issue), W65–70. 10.1093/nar/gks364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider, Z., Subhani, M. M., Farooq, M. A., Ishaq, M., Khalid, M., Khan, R. S., & Ak, N. (2020). In Silico discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 using pharmacophore and molecular docking based virtual screening from ZINC database.
- Imai, Y., Kuba, K., & Penninger, J. M. (2008). The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Experimental Physiology, 93(5), 543–548. 10.1113/expphysiol.2007.040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W., Wang, W., Zhao, X., Zai, J., & Li, X. (2020). Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. Journal of Medical Virology, 92(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalathiya, U., Padariya, M., Mayordomo, M., Lisowska, M., Nicholson, J., Singh, A., Baginski, M., Fahraeus, R., Carragher, N., Ball, K., Haas, J., Daniels, A., Hupp, T. R., & Alfaro, J. A. (2020). Highly Conserved Homotrimer Cavity Formed by the SARS-CoV-2 Spike Glycoprotein: A Novel Binding Site. Journal of Clinical Medicine, 9(5), 1473. 10.3390/jcm9051473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., Collins, R. L., Laricchia, K. M., Ganna, A., Birnbaum, D. P., & Gauthier, L. D. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv, Jan 1:531210. [DOI] [PMC free article] [PubMed]
- Katoh, K., Kuma, K. I., Toh, H., & Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33(2), 511–518. 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid, Z., & Sezerman, O. U. (2020). A Comprehensive Study on Identifying the Structural and Functional SNPs of Human Neuronal Membrane Glycoprotein M6A (GPM6A). Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PubMed] [Google Scholar]
- Khayat, A. S., de Assumpcao, P. P., Khayat, B. C., Araujo, T. M., Batista-Gomes, J. A., Imbiriba, L. C., Ishak, G., de Assumpcao, P. B., Moreira, F. C., & Burbano, R. R. (2020). Ribeiro-dos-Santos AM. ACE2 polymorphisms as potential players in COVID-19 outcome. medRxiv, Jan 1. [DOI] [PMC free article] [PubMed]
- Kouranov, A., Xie, L., de la Cruz, J., Chen, L., Westbrook, J., Bourne, P. E., & Berman, H. M. (2006). The RCSB PDB information portal for structural genomics. Nucleic Acids Research, 34(Database issue), D302–5. 10.1093/nar/gkj120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., Beglov, D., & Vajda, S. (2017). The ClusPro web server for protein–protein docking. Nature Protocols, 12(2), 255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. [DOI] [PubMed]
- Mort, M., Sterne-Weiler, T., Li, B., Ball, E. V., Cooper, D. N., Radivojac, P., Sanford, J. R., & Mooney, S. D. (2014). MutPred Splice: Machine learning-based prediction of exonic variants that disrupt splicing. Genome Biology, 15(1), R19. 10.1186/gb-2014-15-1-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman, H., Bouslama, Z., Brandenburg, J. T., Da Rocha, J., Hamdi, Y., Ghedira, K., Abid, N. S., & Hazelhurst, S. (2020). In silico study of the spike protein from SARS-CoV-2 interaction with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. BioRxiv. [DOI] [PMC free article] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.200844. [DOI] [PubMed] [Google Scholar]
- Pires, D. E., Ascher, D. B., & Blundell, T. L. (2014). mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics (Oxford, England)), 30(3), 335–342. 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva, B., Antipin, Y., & Sander, C. (2011). Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Research, 39(17), e118. 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y., & McCauley, J. (2017). GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance, 22(13). 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. K., Shakya, A., Prasad, S. K., Singh, S., Gurav, N. S., Prasad, R. S., & Gurav, S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium . (2017). UniProt: The universal protein knowledgebase. Nucleic Acids Research, 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P. D., & Kejariwal, A. (2004). Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: Evolutionary evidence for differences in molecular effects. Proceedings of the National Academy of Sciences of the United States of America, 101(43), 15398–15403. 10.1073/pnas.0404380101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venselaar, H., Te Beek, T. A., Kuipers, R. K., Hekkelman, M. L., & Vriend, G. (2010). Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics, 11(1), 548. 10.1186/1471-2105-11-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooster, L., Nicholson, C. J., Sigurslid, H. H., Cardenas, C. L., & Malhotra, R. (2020). Polymorphisms in the ACE2 Locus Associate with Severity of COVID-19 Infection. medRxiv.
- Yan, R., Zhang, Y., Guo, Y., Xia, L., & Zhou, Q. (2020). Structural basis for the recognition of the 2019-nCoV by human ACE2. bioRxiv. [DOI] [PMC free article] [PubMed]
- Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9(1), 40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., & Skolnick, J. (2005). TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Research, 33(7), 2302–2309. 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.