Fig. 1. Possible differential effects of SARS-CoV-2 RNA/proteins on IFN-I and proinflammatory cytokine/chemokine production.
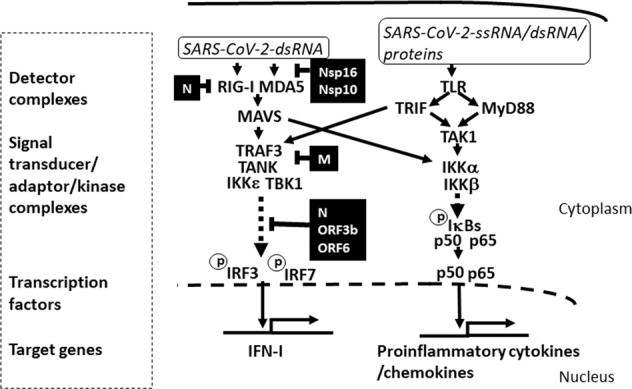
Molecular patterns derived from SARS-CoV-2-associated molecules, such as ssRNA, dsRNA, and viral proteins, bind to host PRRs and trigger the activation of signal transducers and transcription factors that drive the production of IFN-I and proinflammatory cytokines and chemokines. Soon after infection, the engagement of RIG-I and MDA-5 by these molecular patterns induces the activation of IRF3, or IRF7, through MAVS. In addition, viral ssRNA, dsRNA, and proteins can engage TLRs to trigger the MyD88- and TRIF-dependent pathways, primarily leading to the activation of the NF-κB (p50/p65) transcriptional complex. SARS-CoV-2 proteins that inhibit IFN-I production are indicated in black boxes, and the associated blocked pathways are indicated as dashed lines. Note that only the IFN-I production pathway, and not the secretion of proinflammatory cytokines/chemokines, is inhibited by the viral proteins. Proinflammatory cytokine/chemokine production is further activated by the engagement of TLRs by a high viral load.
