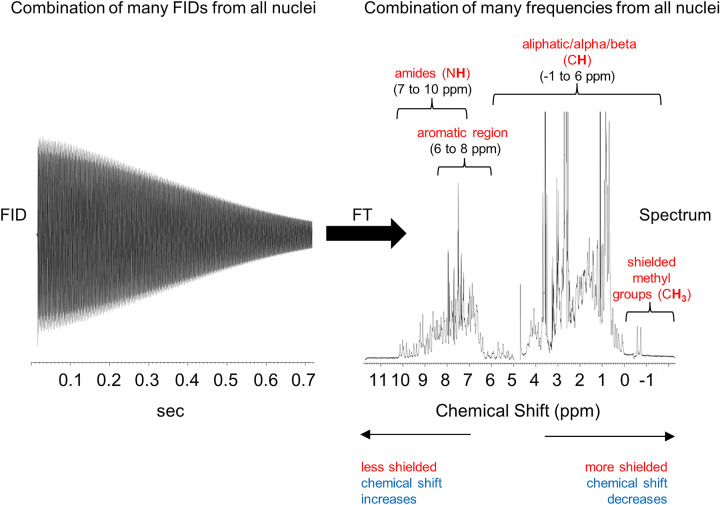
Figure 18. An 1H FID for a protein and its Fourier Transform.
The FID on the left is the sum of FIDs for each different Hydrogen nucleus in the protein. Fourier transformation of this FID creates a set of component frequencies (seen as a peak for each individual FID). Conversion of Larmor frequency (Hz) into chemical shift (ppm) as seen in the 1D 1H NMR spectrum of a protein allows for values to be independent of the magnet strength used. Each peak represents the hydrogen atoms connected to different carbons or nitrogens in the protein. The chemical shifts are different because the 1H nuclei all experience slightly different magnetic environments based on their chemical group and position in the protein and thus their bulk magnetisation vectors rotate at slightly different frequencies. Hydrogens found in common chemical groups (in amides, aromatics, aliphatics, methyl etc.) are indicated above the spectrum. The well-dispersed peaks between 6 and 10 ppm in the backbone amide region indicate that the protein is well folded. It is common to make a higher dimensional spectrum such as the 2D spectrum that plots the chemical shift values for pairs of atoms connected by a covalent bond to better resolve the overlapping signals. Abbreviation: 2D, two dimensional.