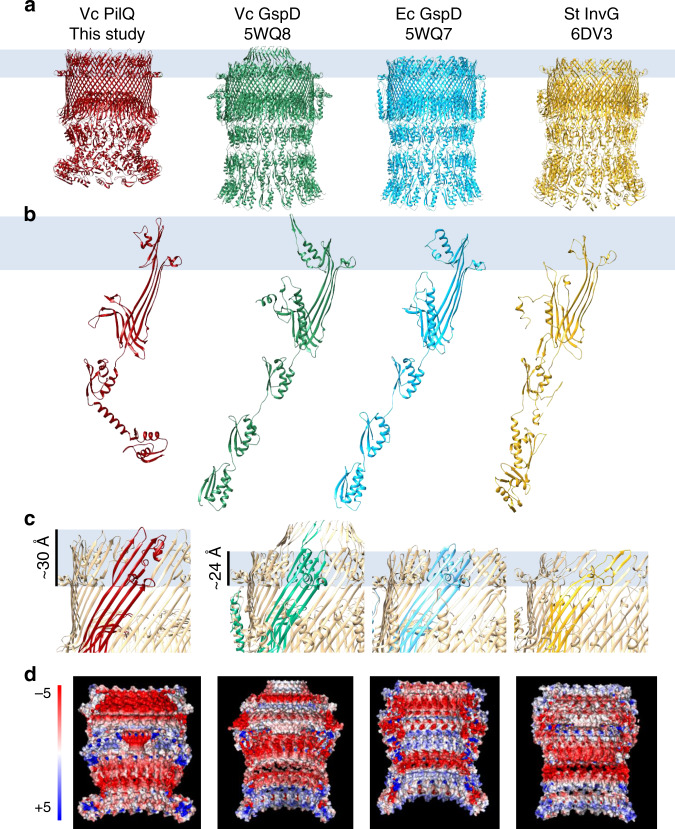
Fig. 2. Comparison of VcPilQ to T2SS and T3SS secretins.
The structures of VcPilQ (this study, dark red), V. cholerae GspD (5WQ8) (green), E. coli K12 GspD (5WQ7) (blue), and S. typhimurium InvG (6DV3) (yellow) are compared55, 61. The putative outer membrane location is depicted as a blue rectangle. The structures are shown as multimers (a) or monomers (b). c The multimer is shown in tan with one subunit colored. The putative outer membrane region is highlighted to show that VcPilQ (left, red) has a ~30 Å membrane spanning distance, while VcGspD, EcGspD, and StInvG have about a 24 Å membrane spanning distance. d Adaptive Poisson–Boltzmann solver was used to calculate the electrostatic potential calculation of each secretin in PyMol142. The inner cavity of each secretin is shown. The scale varies from −5 (red) to +5 (blue) in units of KbT/ec.