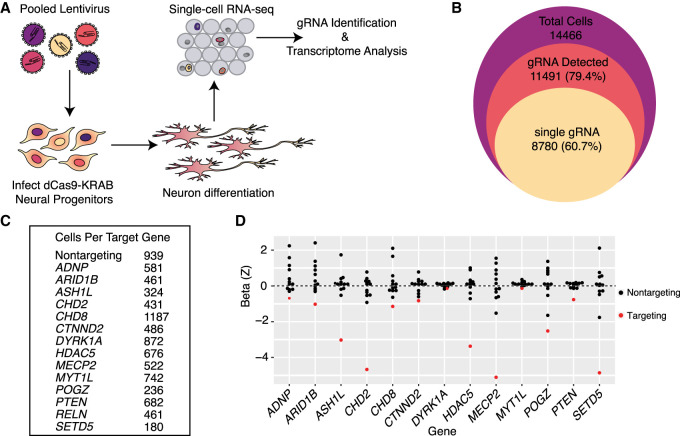
Figure 2.
Single-cell RNA-seq is an efficient readout of multiplexed gene repression in a human model of neuronal differentiation. (A) Schematic of pooled repression of ASD genes in LUHMES. (B) The numbers of total single cells that were collected (purple area), express at least one gRNA (red), and express a single unique gRNA (yellow) show the efficient recovery of gRNAs from single-cell RNA-seq data. (C) Hundreds of cells targeting all 14 genes were recovered, as well as 939 cells with nontargeting gRNAs. (D) Scaled MIMOSCA beta coefficients for targeting and nontargeting guides are shown for each targeted gene. Three targeting gRNAs for each gene are merged. Beta less than zero represents repression. In all cases, targeting gRNAs (red dots) have negative beta coefficients on target gene expression.