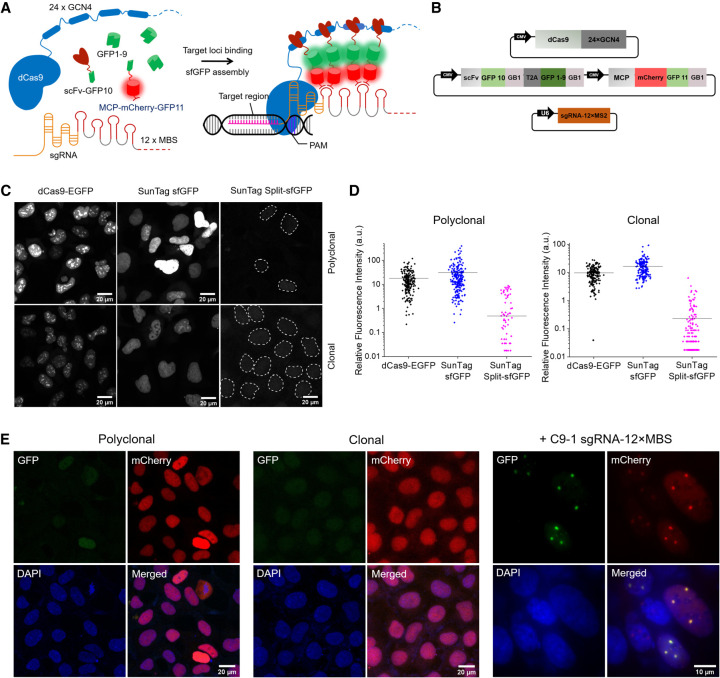
Figure 1.
SunTag split-sfGFP CRISPR system enables background suppression and signal amplification for genome imaging. (A) Schematic design of the CRISPR-dCas9 system, integrated with tripartite sfGFP and the SunTag. dCas9 was fused to 24 × GCN4 peptides to recruit multiple scFv-GFP10 proteins. 12 × MBS motifs were added to the tail of sgRNA to recruit MCP-mCherry-GFP11 proteins. GFP1–9 was expressed separately. (B) Schematic representation of plasmid constructs for the components of the SunTag split-sfGFP CRISPR-dCas9 system. All protein components were expressed under the control of the CMV promoter. sgRNA-12 × MBS was expressed under the control of the U6 promoter. (GB1l) Solubility-enhancing tag, (T2A) self-cleaving peptide. (C) Representative images (GFP channel) of stable AD-293 cell lines transfected with the dCas9-EGFP, SunTag sfGFP, or SunTag split-sfGFP systems (top row) and of homogeneous stable cell lines obtained by colony picking (bottom row), in the absence of sgRNA. Dotted lines indicate nuclear boundaries detected in DAPI channel. (D) Nuclear fluorescence intensity compared between the fluorophore designs in C before and after colony picking (n > 200 cells). (E) Fluorescence images of fixed stable cell lines with the SunTag split-sfGFP CRISPR-dCas9 system in polyclonal and clonal after transient transfection with sgRNA-12 × MBS targeting a pericentromeric region of Chromosome 9, C9-1. mCherry images were acquired to check the expression level of sfGFP fragments.