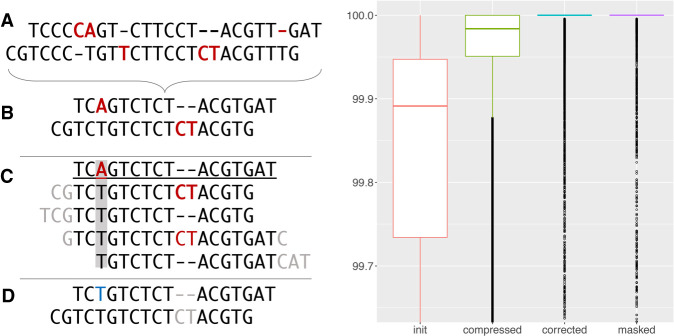
Figure 1.
Impact of HiCanu processing on observed read quality. (A) Two hypothetical reads are shown with sequencing errors highlighted in red. (B) The first step of HiCanu is to compress homopolymers, which obscures homopolymer length errors but retains enough information to accurately distinguish reads from different genomic loci. (C) Overlaps are then computed for the compressed reads, and remaining errors are identified by examining the alignment pileups (gray rectangle). (D) Finally, after correcting the identified errors (blue) and ignoring indels in regions of known systematic error (gray), the resulting overlap is 100% identical. (Right) Sequence identity of reads from a 20-kbp HiFi library measured against the CHM13 Chromosome X reference sequence v0.7 (Miga et al. 2020) after each step of HiCanu processing (Supplemental Note 1). Separate boxplots are shown for initial raw HiFi reads (init), homopolymer-compressed reads (compressed), OEA-corrected reads (corrected), and corrected reads after ignoring differences in microsatellite repeats (masked). The median read identity, indicated by solid segments, increases from <99.9% to 100% (note the plot shows y-range of 99.65%–100%). Supplemental Table S1 also shows how HiCanu processing increases the percentage of perfectly aligned (100% identity) HiFi reads from <1% to >97%.