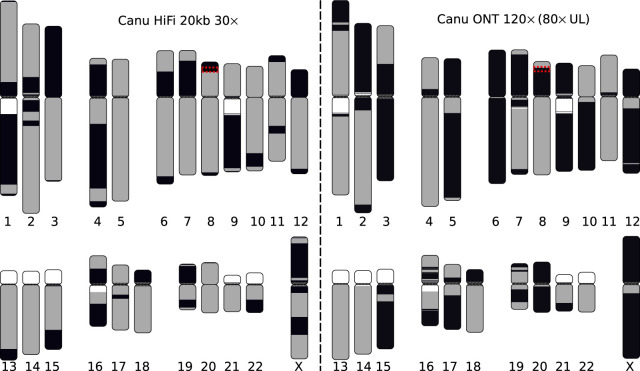
Figure 2.
Visual representation of the most continuous HiFi-based and Nanopore-based assemblies of the CHM13 genome. HiCanu assembly of the 20-kbp HiFi data set (left) and Canu assembly of an ultralong Nanopore data set (right). White regions indicate gaps in the current reference genome, and each gray and black block indicates a continuous contig alignment. Color switches from gray to black represent either the end of a contig or an alignment break. Assemblies were aligned to GRCh38 using MashMap (Jain et al. 2018a), and plots were generated using coloredChromosomes (Böhringer et al. 2002) as previously described (Berlin et al. 2015; Jain et al. 2018b). Note that some chromosomes (e.g., Chr X) are better resolved by the Nanopore assembly owing to the presence of near-perfect repeats. At the same time, chromosomes containing more diverged repeats (e.g., Chr 7 and Chr 16) are better resolved by the HiFi assembly. We note that some gaps in the HiFi assembly are caused by sequence-specific biases of current HiFi sequencing protocols (Supplemental Note 4). The red box highlights the defensin beta gene family on Chromosome 8p23.1 which is split in both assemblies and detailed in Figure 4.