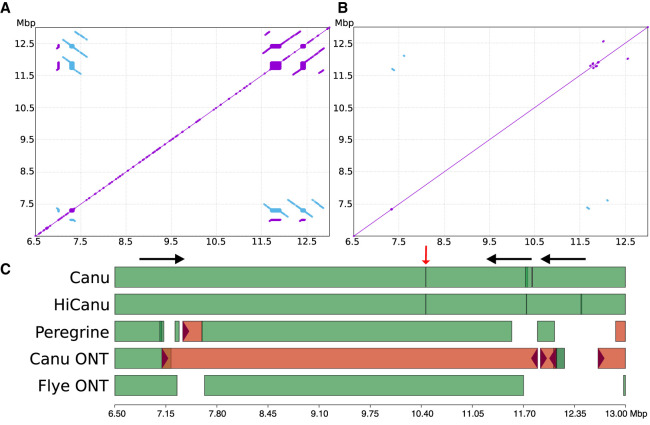
Figure 4.
Chr 8 defensin beta cluster repeat structure and assembly comparison. (Top) NUCmer self-alignment dot plots (Kurtz et al. 2004) of the CHM13 reference defensin beta cluster at different alignment stringencies (Methods): (A) >7 kbp repeats at 98% identity. (B) >7 kbp repeats at 99.9% identity. Purple/blue indicates same/reverse strand matches. (C) Icarus (Mikheenko et al. 2016) visualization of contig alignments from both HiFi-based (Canu, HiCanu, Peregrine) and ultralong Nanopore-based assemblies (Canu ONT and Flye ONT) (Kolmogorov et al. 2019) produced by QUAST (Gurevich et al. 2013). White space in the alignment figure indicates the assembly was fragmented into short contigs (<50 kbp). Red color indicates misassembled contigs. The HiCanu assembly breaks at two of three SD instances that share high sequence similarity (black arrows) and at a region of systematic HiFi coverage depletion (red arrow).