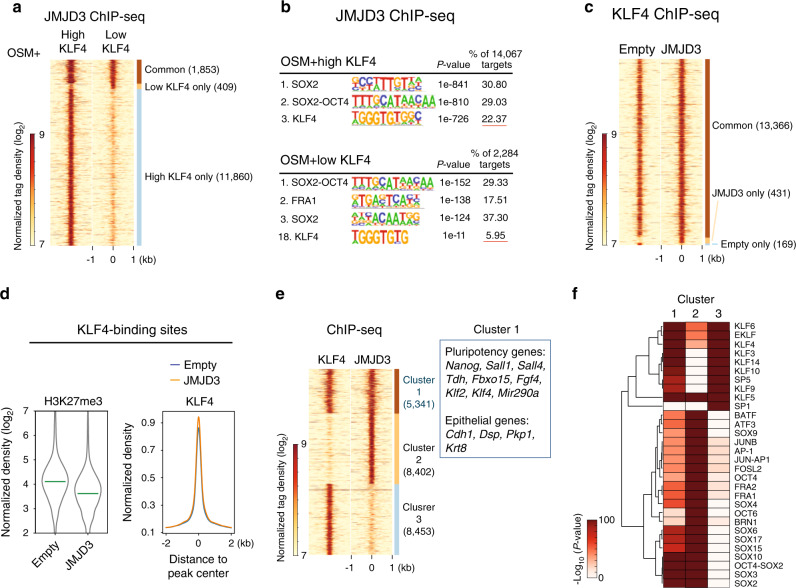
Fig. 5. The genome-wide cooperation of KLF4 and JMJD3 during reprogramming.
a Tag density heatmaps of JMJD3 peaks in P2 MEFs transduced with OSM + JMJD3 and low or high level of KLF4 in medium with Vc at day 5. Peaks were merged into the same genomic loci if the summits were within 400 bp, and the resulting merged peaks were allocated to clusters as indicated: common between high and low (Common), low KLF4-specific (Low KLF4 only), and High KLF4-specific (High KLF4 only). b De novo motif discovery by HOMER at JMJD3 binding sites. c Tag density heatmaps of KLF4 binding in P2 MEFs transduced with OSKM and Empty or JMJD3 in medium with Vc at day 5. Peaks were merged into the same genomic loci if the summits were within 400 bp and the resulting merged peaks were allocated to clusters as indicated: common between Empty and JMJD3 (Common), JMJD3-specific (JMJD3 only), and Empty-specific (Empty only). d Violin plots of the levels of H3K27me3 at KLF4-binding sites (left panel) and the pileup tag density plots of KLF4-binding intensity (right panel) with Empty or JMJD3. e Tag density heatmaps of JMJD3 or KLF4 peaks. Peaks were merged into the same genomic loci if the summits were within 400 bp, and the resulting merged peaks were allocated to the indicated clusters. Cluster 1: common between JMJD3 and KLF4; Cluster 2: JMJD3-specific; Cluster 3: KLF4-specific. Representative pluripotency and epithelial genes co-bound by KLF4 and JMJD3 in Cluster 1 are listed. f Enrichment of specific transcription factor motifs (from HOMER) in each cluster. Data were grouped based on a Euclidean distance matrix and complete linkage.