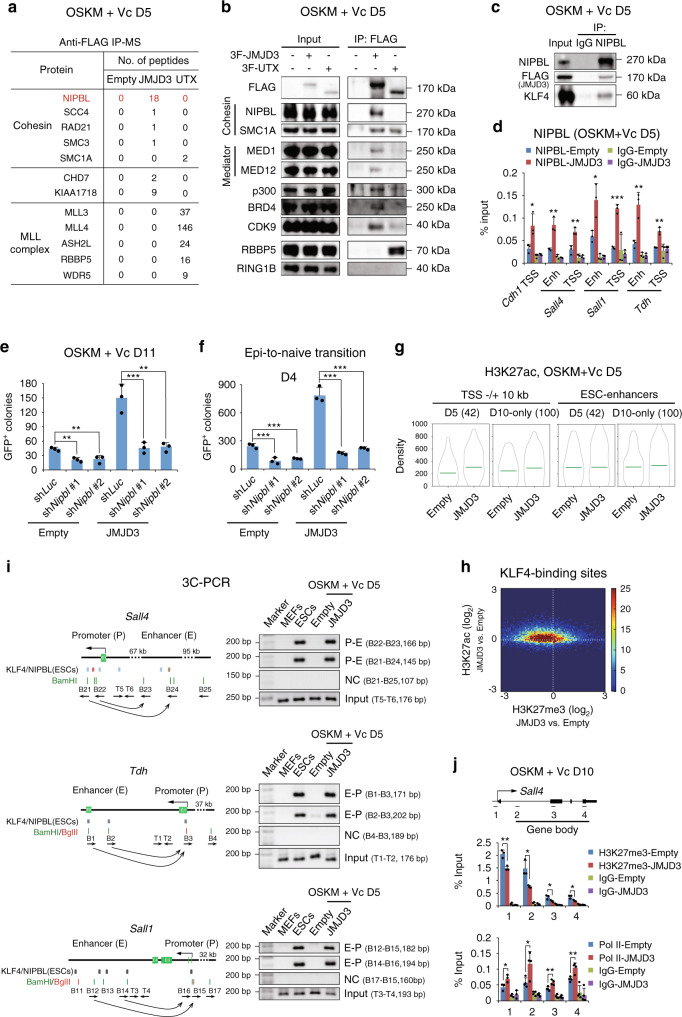
Fig. 6. JMJD3 facilitates enhancer–promoter looping and transcriptional activation.
a, b List of proteins identified by MS a and western blotting with indicated antibodies b following immunoprecipitation with anti-FLAG beads in nuclear extracts from OSKM-reprogrammed P2 MEFs with FLAG-tagged JMJD3 or UTX. RING1B is the negative control. c Immunoprecipitation with NIPBL antibody in nuclear extracts from OSKM-reprogrammed P2 MEFs with FLAG-tagged JMJD3, followed by western blotting with the indicated antibodies. d ChIP-qPCR for NIPBL at the enhancer/TSS of the indicated genes in OSKM-reprogrammed P2 MEFs with Empty or JMJD3. e Number of GFP+ colonies in OSKM-reprogrammed P2 MEFs with Empty or JMJD3 plus shLuc or shNipbl. f Number of GFP+ colonies in OG2 EpiSCs transduced with KLF4 and Empty or JMJD3 plus shLuc or shNipbl. g Violin plots of H3K27ac levels in OSKM-reprogrammed P2 MEFs with Empty or JMJD3 at TSS/enhancers of PSC-enriched genes upregulated upon JMJD3 OE at day 5 or only at day 10. h Correlation of H3K27me3 and H3K27ac changes at KLF4-binding sites by JMJD3 OE in OSKM-reprogrammed P2 MEFs with Empty or JMJD3 in medium with Vc at day 5, shown as a 2D density histogram where each point represents a KLF4-binding site. i Schematic (left panels) and 3C-PCR (right panels) of enhancer–promoter looping between the indicated sites. Arrows under restriction enzyme sites mark the orientation of the primers. j ChIP-qPCR for H3K27me3 and Pol II at the indicated loci of Sall4 in OSKM-reprogrammed P2 MEFs with Empty or JMJD3. Error bars represent the s.e.m. Data are presented as mean ± s.e.m. from n = 3 d–f, j biologically independent experiments. Statistical analyses were performed using a two-tailed unpaired Student’s t-test d, j or one-way ANOVA followed by a Dunnett multiple comparison test e, f (*P < 0.05; **P < 0.01; ***P < 0.001). P values: 0.0343, 0.0059, 0.0059, 0.0228, 0.0001, 0.0062, 0.0019 d; 0.0049, 0.0053, 0.0009, 0.0010 e; 0.0004, 0.0007, 0.0001, 0.0001 f; 0.0054, 0.0193, 0.0364, 0.0161 (H3K27me3), 0.0419, 0.0288, 0.0074, 0.0063 (Pol II) j. Experiments were repeated independently three times b or twice c, j with similar results. Source data are provided as a Source Data file.