Abstract
Sickle Cell Disease (SCD) is a hereditary disorder of red blood cells in humans. Complications such as pain, stroke, and organ failure occur in SCD as malformed, sickled red blood cells passing through small blood vessels get trapped. Particularly, acute pain is known to be the primary symptom of SCD. The insidious and subjective nature of SCD pain leads to challenges in pain assessment among Medical Practitioners (MPs). Thus, accurate identification of markers of pain in patients with SCD is crucial for pain management. Classifying clinical notes of patients with SCD based on their pain level enables MPs to give appropriate treatment. We propose a binary classification model to predict pain relevance of clinical notes and a multiclass classification model to predict pain level. While our four binary machine learning (ML) classifiers are comparable in their performance, Decision Trees had the best performance for the multiclass classification task achieving 0.70 in F-measure. Our results show the potential clinical text analysis and machine learning offer to pain management in sickle cell patients.
Index Terms—: Sickle Cell Disease, Pain Management, Text Mining, Machine Learning
I. Introduction
Sickle cell disease (SCD) affects nearly 100,000 people in the US1 and is an inherited red blood cell disorder. Common complications of SCD include acute pain, organ failure, and early death [1]. Acute pain arises in patients when blood vessels are obstructed by sickle-shaped red blood cells mitigating the flow of oxygen, a phenomenon called vaso-occlusive crisis. Further, pain is the leading cause of hospitalizations and emergency department admissions for patients with SCD. The numerous health care visits lead to a massive amount of electronic health record (EHR) data, which can be leveraged to investigate the relationships between SCD and pain. Since SCD is associated with several complications, it is important to identify clinical notes with signs of pain from those without pain. It is equally important to gauge changes in pain for proper treatment.
Due to their noisy nature, analyzing clinical notes is a challenging task. In this study, we propose techniques employing natural language processing, text mining and machine learning to predict pain relevance and pain change from SCD clinical notes. We build two kinds of models: 1) A binary classification model for classifying clinical notes into pain relevant or pain irrelevant; and 2) A multiclass classification model for classifying the pain relevant clinical notes into i) pain increase, ii) pain uncertain, iii) pain unchanged, and iv) pain decrease. We experiment with Logistic Regression, Decision Trees, Random Forest, and Feed Forward Neural Network (FFNN) for both the binary and multiclass classification tasks. For the multiclass classification task, we conduct ordinal classification as the task is to predict pain change levels ranging from pain increase to pain decrease. We evaluate the performance of our ordinal classification model using graded evaluation metrics proposed in [2].
II. Related Work
There is an increasing body of work assessing complications within SCD. Mohammed et al. [1] developed an ML model to predict early onset organ failure using physiological data of patients with SCD. They used five physiologic markers as features to build a model using a random forest classifier, achieving the best mean accuracy in predicting organ failure within six hours before the incident. Jonassaint et al. [3] developed a mobile app to monitor signals such as clinical symptoms, pain intensity, location and perceived severity to actively monitor pain in patients with SCD. Yang et al. [4] employed ML techniques to predict pain from objective vital signs shedding light on how objective measures could be used for predicting pain.
Past work on predicting pain or other comorbidities of SCD, has thus, relied on features such as physiological data to assess pain for a patient with SCD. In this study, we employ purely textual data to assess the prevalence of pain in patients and whether pain increases, decreases or stays constant.
There have been studies on clinical text analysis for other classification tasks. Wang et al. [5] conducted smoking status and proximal femur fracture classification using the i2b2 2006 dataset. Chodey et al. [6] used ML techniques for named entity recognition and normalization tasks. Elhadad et al. [7] conducted clinical disorder identification using named entity recognition and template slot filling from the ShARe corpus (Pradhan et al., 2015) [8]. Similarly, clinical text can be used for predicting the prevalence and degree of pain in sickle cell patients as it has a rich set of indicators for pain.
III. Data Collection
Our dataset consists of 424 clinical notes of 40 patients collected by Duke University Medical Center over two years (2017 – 2019). The clinical notes are jointly annotated by two co-author domain experts. There are two rounds of annotation conducted on the dataset. In the first round, the clinical notes were annotated as relevant to pain or irrelevant to pain. In the second round, the relevant to pain clinical notes were annotated to reflect pain change. Figure-1 shows the size of our dataset based on pain relevance and pain change. As shown, our dataset is mainly composed of pain relevant clinical notes. Among the pain relevant clinical notes, clinical notes labeled pain decrease for the pain change class outnumber the rest. Sample pain relevant and pain irrelevant notes are shown in Table-I.
Fig. 1.
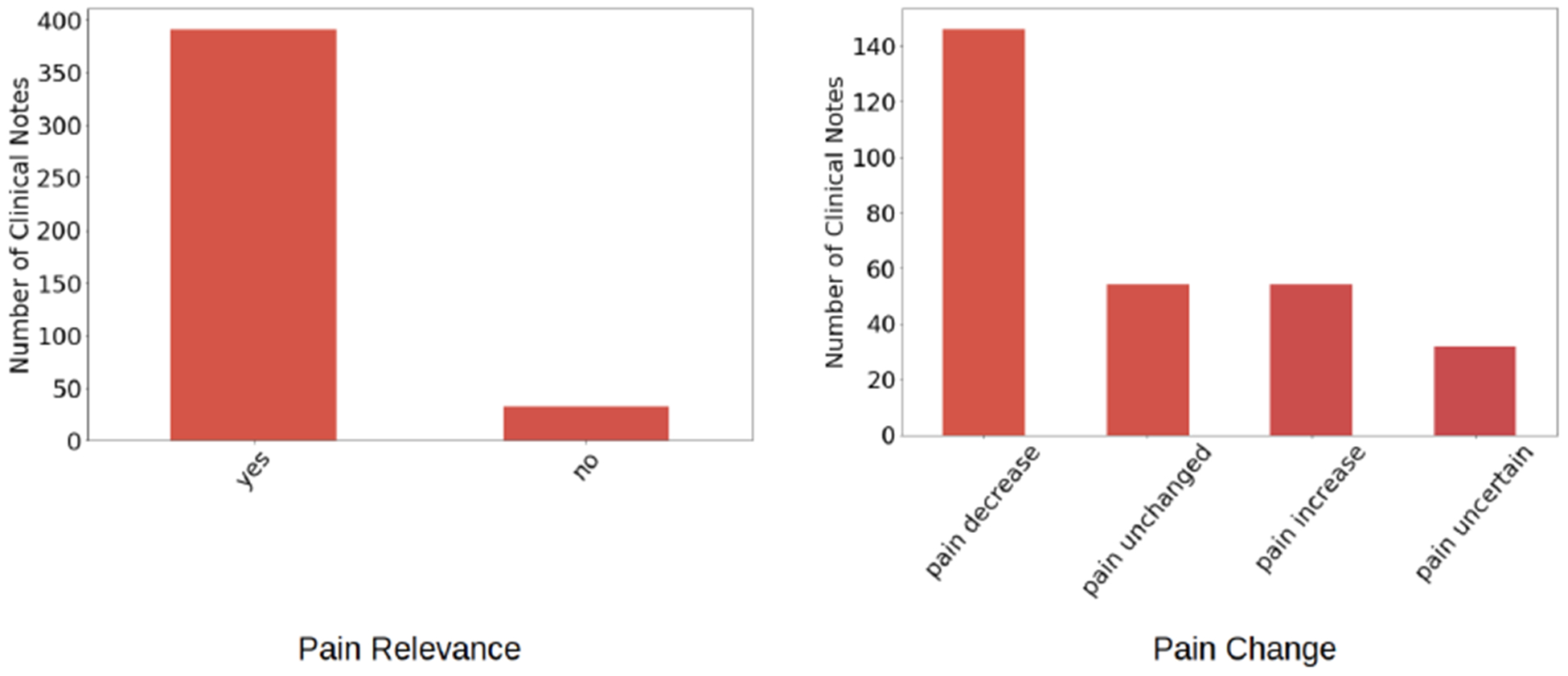
Statistics of dataset for Pain Relevance and Pain Change classes
TABLE I.
Sample Clinical notes
| Pain Relevance | Sample Clinical Note |
|---|---|
| YES | Patient pain increased from 8/10 to 9/10 in chest. |
| NO | Discharge home |
Our dataset is highly imbalanced, particularly, among the pain relevance classes. There are significantly higher instances of clinical notes labeled pain relevant than pain irrelevant. To address this imbalance in our dataset, we employed a technique called Synthetic Minority Over-sampling TEchnique (SMOTE) [9] for both classification tasks.
We preprocessed our dataset by removing stop words as well as punctuations, and performed lemmatization.
IV. Methods
The clinical notes are labeled by co-author domain experts based on their pain relevance and pain change indicators. The pain change labels use a scale akin to the Likert scale from severe to mild. Our pipeline (Figure-2) consists of data collection, data preprocessing, linguistic/topical analysis, feature extraction, feature selection, model creation, and evaluation. We use linguistic and topical features to build our models. While linguistic analysis is used to extract salient features, topical features are used to mine latent features. We performed two sets of experiments: 1) Binary Classification for pain relevance classification, and 2) Multiclass Classification for pain change classification.
Fig. 2.
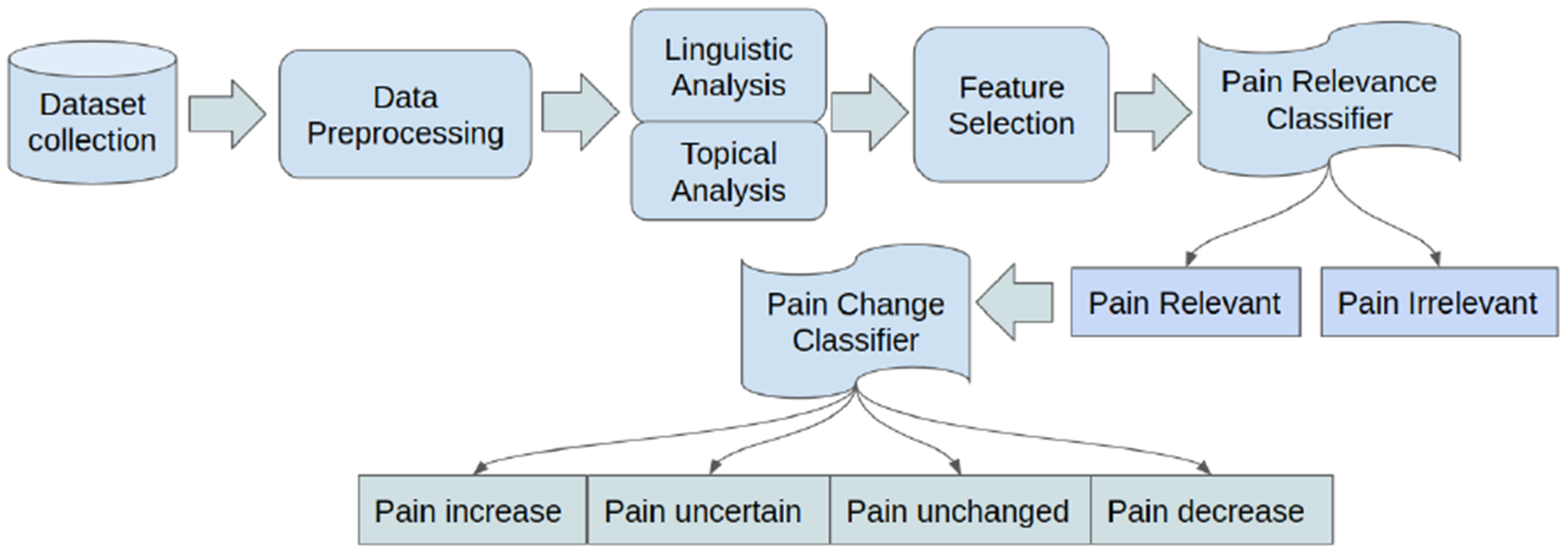
Sickle Cell Disease Pain Classification Pipeline
A. Linguistic Analysis
To infer salient features in our dataset, we performed linguistic analysis. We generated n-grams for pain-relevant and pain-irrelevant clinical notes and clinical notes labeled pain increase, pain uncertain, pain unchanged, or pain decrease. In our n-grams analysis, we observe there are unigrams and bigrams that are common to different classes (e.g., common to pain relevant and pain irrelevant). Similarly, there are unigrams and bigrams that are exclusive to a given class. Table-II shows the top 10 unigrams selected using χ2 feature selection for our dataset based on the classes of interest.
TABLE II.
Top 10 Unigrams
| Pain Relevant (Exclusive) | Pain Irrelevant (Exclusive) | Pain Relevant AND Pain Irrelevant |
|---|---|---|
| emar, intervention, increase, dose, expressions, chest, regimen, alteration, toradol, medication | home, wheelchair, chc, fatigue, bedside, parent, discharge, warm, relief, mother | pain, pca, plan, develop, control, altered, patient, level, comfort, manage |
B. Topical Analysis
While n-grams analysis uncovers explicit language features in the clinical notes, it is equally important to uncover the hidden features characterizing the topical distribution. We adopt the Latent Dirichlet Allocation (LDA) [10] for unraveling these latent features. We train an LDA model using our entire corpus.
To determine the optimal number of topics for a given class of clinical notes (e.g., pain relevant notes), we computed coherence scores [11]. The higher the coherence score for a given number of topics, the more intepretable the topics are (see Figure-3). We set the number of words characterizing a given topic to eight. These are words with the highest scores in the topic distribution. We found the human-interpretable optimal number of topics for each of the classes of the clinical notes in our dataset to be two. This is interpreted as each class of the clinical notes is a mixture of two topics. Table-III shows words for the two topics for pain relevant and pain irrelevant clinical notes. As can be seen in the table, pain relevant notes can be interpreted to have mainly the topic of pain control, while pain irrelevant notes to have primarily the topic of home care. Similarly, Table-IV shows the distribution of words for the topics for each of the pain change classes (underscored words are exclusive to the corresponding class for Topic-1). Further, pain appears in each of the topics for pain change classes and, as a result, is not discriminative. While a common word such as pain in the topic distribution can be considered as a stop word and not helpful for pain change classification, we did not remove it since pain helps with interpretation of a given topic regardless of other topics.
Fig. 3.
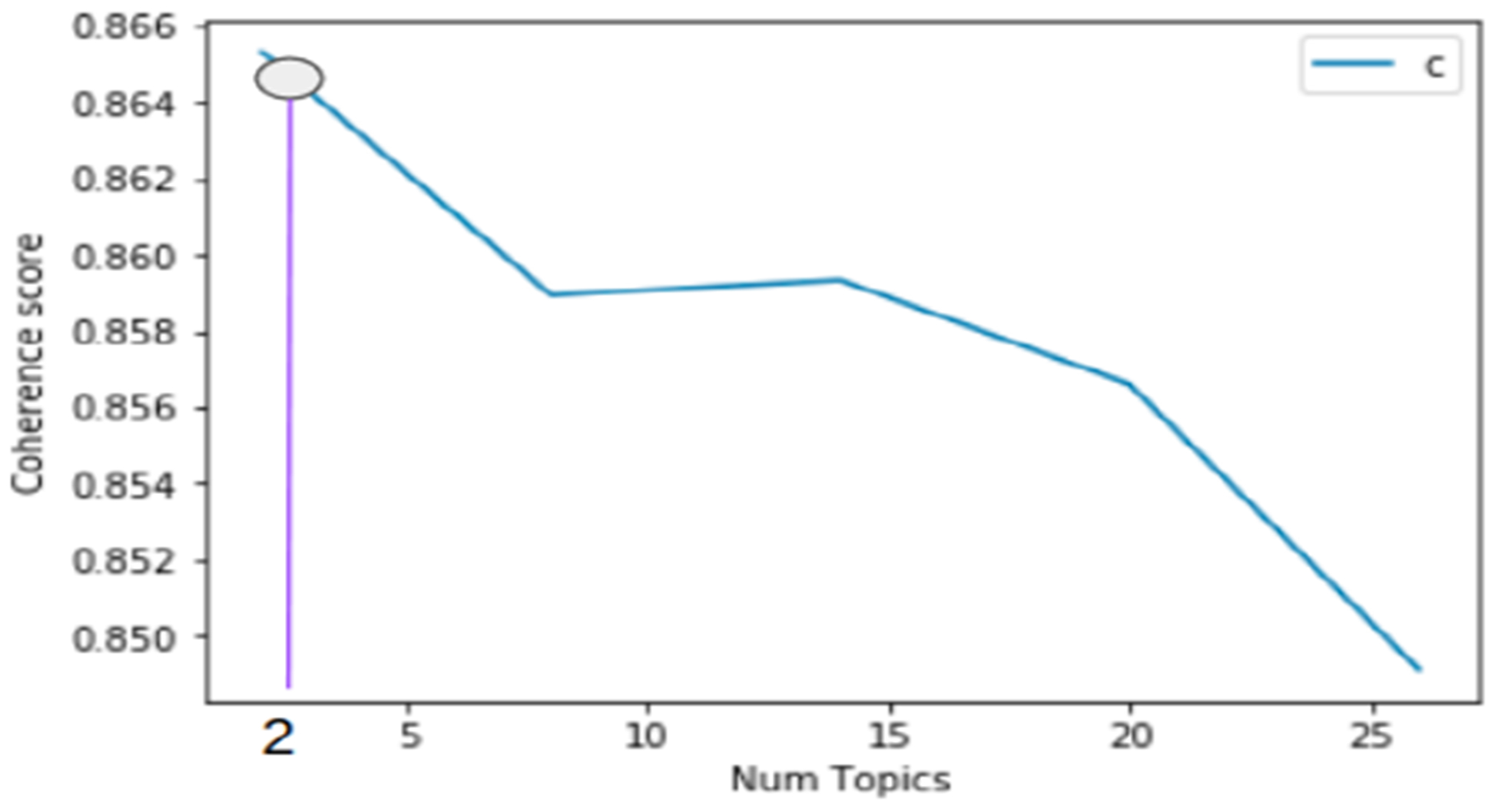
Coherence Scores vs Number of Topics
TABLE III.
Topic distribution based on pain relevance
| Pain Relevance | Most Prevalent Words in Topic-1 | Most Prevalent Words in Topic-2 |
|---|---|---|
| YES | progress, pain, improve, decrease, knowledge, control | patient, pain, medication, knowledge, goal, state |
| NO | note, admission, discharge, patient, home, ability | pain, goal, admission, outcome, relief, continue |
TABLE IV.
Topic distribution based on pain change
| Pain Change | Most Prevalent Words in Topic-1 | Most Prevalent Words in Topic-2 |
|---|---|---|
| Pain increase | pain, progress, medication, management, patient, schedule, pca, intervention | pain, patient, give, goal, intervention, dose, button, plan |
| Pain uncertain | pain, patient, goal, continue, plan, improve, decrease, develop | outcome, pain, problem, knowledge, regimen, deficit, carry, method |
| Pain unchanged | pain, progress, level, control, develop, plan, regimen, pca | patient, pain, remain, well, demand, plan, level, manage |
| Pain decrease | pain, progress, patient, decrease, plan, regimen, satisfy, alter | pain, patient, improve, satisfy, control, decrease, manage, ability |
C. Classification
The language and topical analyses results are used as features in building the ML models. Our classification task consists of two sub-classification tasks: 1) pain relevance classification; 2) pain change classification, each with its own sets of features. The pain relevance classifier classifies clinical notes into pain-relevant and pain-irrelevant. The pain change classifier is used to classify the pain-relevant clinical notes into 1) pain increase, 2) pain uncertain, 3) pain unchanged, and 4) pain decrease. We trained and evaluated various ML models for each classification task. We used a combination of different linguistic and topical features to train our models. Since linguistic and topical features are generated using independent underlying techniques, which make them orthogonal, concatenation operation is used to combine their representations. We split our dataset into 80% training and 20% testing sets and built logistic regression, decision trees, random forests, and FFNN for both classification tasks. Table-V shows the results of the pain relevance classifier while Table-VI shows pain change classification results. For the ordinal classification, we considered the following order in the severity of pain change from high to low: pain increase, pain uncertain, pain unchanged, pain decrease.
TABLE V.
Pain Relevance Classification
| Model | Feature | Precision | Recall | F-measure |
|---|---|---|---|---|
| Logistic Regression | Linguistic | 0.94 | 0.93 | 0.94 |
| Topical | 0.98 | 0.86 | 0.91 | |
| Linguistic + Topical | 0.95 | 0.95 | 0.95 | |
| Decision Trees | Linguistic | 0.95 | 0.95 | 0.95 |
| Topical | 0.98 | 0.98 | 0.98 | |
| Linguistic + Topical | 0.98 | 0.98 | 0.98 | |
| Random Forest | Linguistic | 0.90 | 0.95 | 0.92 |
| Topical | 0.95 | 0.98 | 0.98 | |
| Linguistic + Topical | 0.90 | 0.95 | 0.93 | |
| FFNN | Linguistic | 0.94 | 0.94 | 0.94 |
| Topical | 0.98 | 0.98 | 0.98 | |
| Linguistic + Topical | 0.96 | 0.96 | 0.94 |
TABLE VI.
Pain Change Classification
| Model | Feature | Precision | Recall | F-measure |
|---|---|---|---|---|
| Logistic Regression | Linguistic | 0.75 | 0.56 | 0.63 |
| Topical | 0.50 | 0.55 | 0.52 | |
| Linguistic + Topical | 0.76 | 0.58 | 0.66 | |
| Decision Trees | Linguistic | 0.76 | 0.59 | 0.67 |
| Topical | 0.73 | 0.65 | 0.68 | |
| Linguistic + Topical | 0.74 | 0.68 | 0.70 | |
| Random Forest | Linguistic | 0.74 | 0.49 | 0.59 |
| Topical | 0.94 | 0.52 | 0.66 | |
| Linguistic + Topical | 0.81 | 0.46 | 0.59 | |
| FFNN | Linguistic | 0.71 | 0.59 | 0.65 |
| Topical | 0.73 | 0.65 | 0.68 | |
| Linguistic + Topical | 0.83 | 0.51 | 0.63 |
V. Discussion
For pain relevance classification, the four models have similar performance. For pain change classification, however, we see a significant difference in performance across the various combinations of features and models. Decision trees with linguistic and topical features achieve the best performance in F-measure. While random forest, and FFNN offer better precision, each, than decision tree, they suffer on Recall, and therefore on F-measure. Further, most models perform better when trained on topical features than pure linguistic features. A combination of topical and linguistic features usually offers the best model performance. Thus, latent features obtained using LDA enable an ML model to perform better.
Evaluation of the multiclass classification task is conducted using the techniques used by Gaur et al. [2] where a model is penalized based on how much it deviates from the true label for an instance. Formally, the count of true positives is incremented when the true label and predicted label of an instance are the same. Similarly, false positives’ count gets incremented by an amount equal to the gap between a predicted label and true label (when predicted label is higher than true label). False negatives’ count is incremented by the difference between the predicted label and true label (when predicted label is lower than true label). Precision, and recall are then computed following the implementations defined in ML libraries2 using the count of true positives, false positives, and false negatives. Finally, F-measure is defined as the harmonic mean of precision and recall.
While we achieved scores on the order of 0.9 for pain relevance classification, the best we achieved for pain change classification was 0.7. This is because there is more disparity in linguistic and topical features between pain relevant and pain irrelevant notes than there is among the four pain change classes. since the price of false negatives is higher than false positives in a clinical setting, we favor decision trees with n-grams and topics used as features as they achieve the best Recall and F-measure, albeit they lose to other models on Precision. Thus, identification of pain relevant notes with 0.98 F-measure followed by a 0.70 F-measure on determining pain change is impressive. We believe our model can be used by MPs for sCD-induced pain mitigation.
VI. Conclusion and Future Work
In this study, we conducted a series of analyses and experiments to leverage the power of natural language processing and ML to predict pain relevance and pain change from clinical text. specifically, we used a combination of linguistic and topical features to build different models and compared their performance. Results show decision tree followed by feed forward neural network as the most promising models.
In future work, we plan to collect additional clinical notes and use unsupervised, and deep learning techniques for predicting pain. Further, we look forward to fusing different modalities of sickle cell data for better modeling of pain or different physiological manifestations of SCD.
Acknowledgments
This paper is based on work supported by the National Institutes of Health under Grant no 1R01AT010413-01. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NIH. The experimental procedures involving human subjects described in this paper were approved by the Institutional Review Board.
Footnotes
References
- [1].Mohammed A, Podila PS, Davis RL, Ataga KI, Hankins JS, and Kamaleswaran R, “Machine learning predicts early-onset acute organ failure in critically ill patients with sickle cell disease,” bioRxiv, p. 614941, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gaur M, Alambo A, Sain JP, Kursuncu U, Thirunarayan K, Kavuluru R, Sheth A, Welton R, and Pathak J, “Knowledge-aware assessment of severity of suicide risk for early intervention,” in The World Wide Web Conference ACM, 2019, pp. 514–525. [Google Scholar]
- [3].Jonassaint CR, Shah N, Jonassaint J, and De Castro L, “Usability and feasibility of an mhealth intervention for monitoring and managing pain symptoms in sickle cell disease: the sickle cell disease mobile application to record symptoms via technology (smart),” Hemoglobin, vol. 39, no. 3, pp. 162–168, 2015. [DOI] [PubMed] [Google Scholar]
- [4].Yang F, Banerjee T, Narine K, and Shah N, “Improving pain management in patients with sickle cell disease from physiological measures using machine learning techniques,” Smart Health, vol. 7, pp. 48–59, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Y, Sohn S, Liu S, Shen F, Wang L, Atkinson EJ, Amin S, and Liu H, “A clinical text classification paradigm using weak supervision and deep representation,” BMC medical informatics and decision making, vol. 19, no. 1, p. 1, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chodey KP and Hu G, “Clinical text analysis using machine learning methods,” in 2016 IEEE/ACIS 15th International Conference on Computer and Information Science (ICIS) IEEE, 2016, pp. 1–6. [Google Scholar]
- [7].Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, and Savova G, “Semeval-2015 task 14: Analysis of clinical text,” in Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015), 2015, pp. 303–310. [Google Scholar]
- [8].Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, Suominen H, Chapman WW, and Savova G, “Evaluating the state of the art in disorder recognition and normalization of the clinical narrative,” Journal of the American Medical Informatics Association, vol. 22, no. 1, pp. 143–154, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chawla NV, Bowyer KW, Hall LO, and Kegelmeyer WP, “Smote: synthetic minority over-sampling technique,” Journal of artificial intelligence research, vol. 16, pp. 321–357, 2002. [Google Scholar]
- [10].Blei DM, Ng AY, and Jordan MI, “Latent dirichlet allocation,” Journal of machine Learning research, vol. 3, no. Jan, pp. 993–1022, 2003. [Google Scholar]
- [11].Stevens K, Kegelmeyer P, Andrzejewski D, and Buttler D, “Exploring topic coherence over many models and many topics,” in Proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning Association for Computational Linguistics, 2012, pp. 952–961. [Google Scholar]


