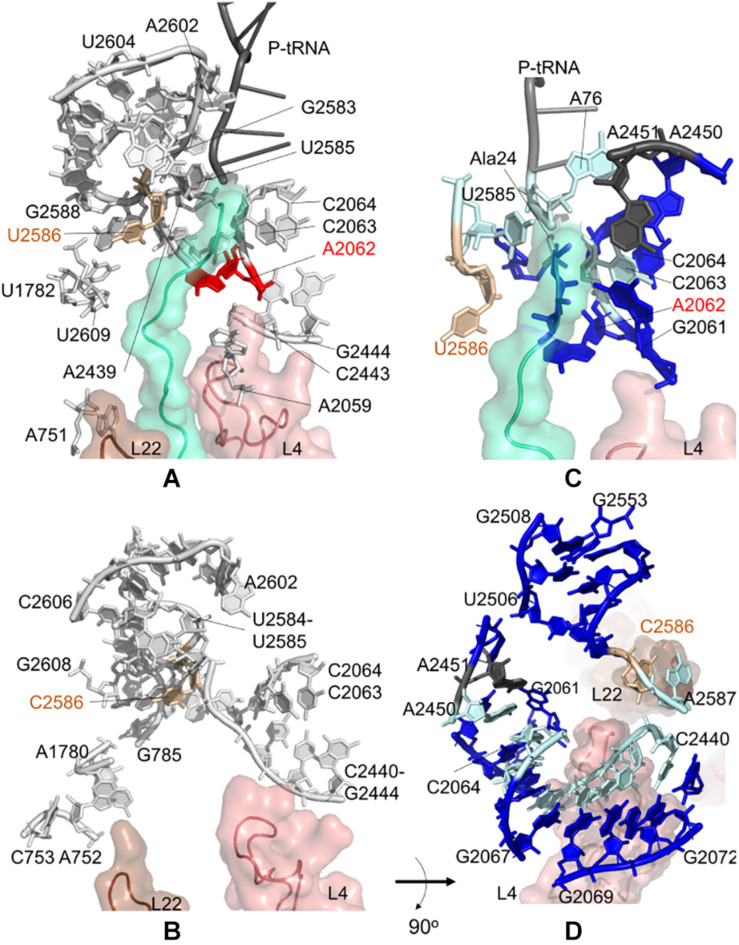
FIGURE 3.
Nucleotides/residues from the sensitivity profile of U2585–U2586 based on CGMD simulations of (A) E. coli ribosomal complex with PDB ID 4v5h, and (B) T. thermophilus with PDB ID 4v5d. Nucleotides forming the k-shortest pathways on (C) E. coli large subunit with PDB ID 4v5h, and (D) T. thermophilus conformers generated by ClustENM using crystal structure with PDB ID 4v9m. Coloring is as in Figure 2.