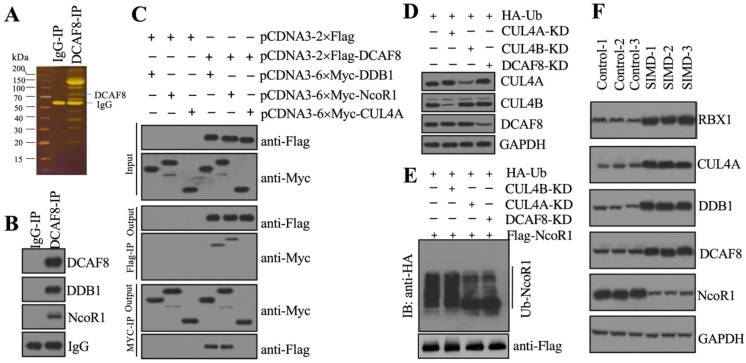
Figure 3.
CRL4ADCAF8 recognized NcoR1 as a substrate in vivo. (A) DCAF8-associated proteins in vivo. Equal weights of hear tissues (n=3) were mixed together and lysed in RIPA buffer, followed by IP with IgG (negative control) and anti-DCAF8-coupled protein A beads. The purified DCAF8-associated protein complex was stained by a silver staining kit. The DCAF8 and IgG bands were indicated. (B) DCAF8 could pull down NcoR1 in vivo. The IP products used in (A) were applied to immunoblots to detect the protein levels of DCAF8, DDB1 and DCAF8. IgG was used as a loading control. (C) DCAF8 could pull down both DDB1 and NcoR1 in vitro. Cells were cotransfected with the plasmid combinations as indicated in the figure, followed by co-IP analyses using anti-Myc-agarose or anti-Flag-agarose beads. The input and output proteins were used to detect protein levels with anti-Flag and anti-Myc antibodies. (D) The protein levels of CUL4A/4B and DCAF8. The Control, CUL4A-KD, CUL4B-KD and DCAF8-KD cells coexpressing pcDNA3-2×Flag-NcoR1 and HA-ubiquitin were used for immunoblotting to examine the protein levels of CUL4A, CUL4B and DCAF8. GAPDH was used as a loading control. (E) The in vivo ubiquitination of NcoR1. Cells used in (D) were immunoprecipitated with an anti-Flag-agarose, and the ubiquitination of NcoR1 was detected using an anti-HA antibody. FlagNcoR1 was used as a loading control. (F) The protein levels of CRL4ADCAF8 components and NcoR1 in SIMD heart tissues. Total cell extracts from three independent heart tissues of control and SIMD mice were used for western blotting to examine the protein levels of RBX1, CUL4A, DDB1, DCAF8 and NcoR1. GAPDH was used as a loading control.