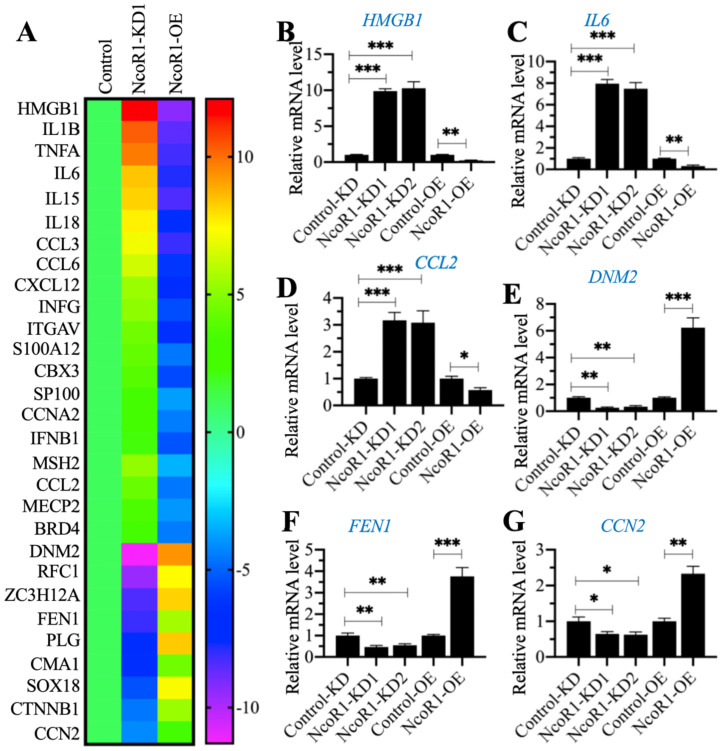
Figure 4.
The identification and verification of NcoR1-dependent genes. (A) The heat map of differentially expressed genes dependent on NcoR1. Total RNA samples from RAW246.7 (Control), NcoR1-KD1 and NcoR1-OE cells were used for microarray analysis. The aberrantly expressed genes that showed converse expression patterns in NcoR1-KD1 and NcoR1-OE cells were selected and presented in a heat map. (B-G) RT-qPCR results. Total RNA samples isolated from Control-KD, NcoR1-KD (#1 and #2), Control-OE, and NcoR1-OE cells were used to detect the mRNA levels of six genes, HMGB1 (B), IL6 (C), CCL2 (D), DNM2 (E), FEN1 (F), and CCN2 (G). * P < 0.05, ** P < 0.01 and *** P < 0.001.