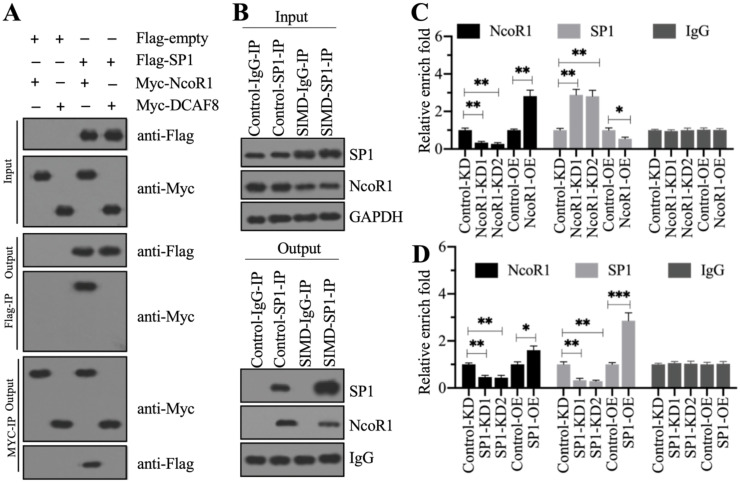
Figure 6.
NcoR1 directly interacted with SP1 to suppress its occupancy on the promoter of HMGB1. (A) SP1 could pull down NcoR1 in vitro. Cells were cotransfected with the plasmid combinations as indicated in the figure, followed by co-IP analyses using anti-Myc-agarose or anti-Flag-agarose beads. The input and output proteins were used to detect protein levels with anti-Flag and anti-Myc antibodies. (B) SP1 could pull down NcoR1 in vivo. Equal weights of heart tissues from three control or SIMD mice were mixed and then lysed in RIPA buffer to isolate total proteins. Total cell extracts were subjected to IP assays using IgG and anti-SP1-coupled protein A beads. The input and output proteins were used for immunoblotting to examine the protein levels of SP1 and NcoR1. GAPDH and IgG were the loading controls of input and output, respectively. (C) The relative occupancies of NcoR1 and SP1 on the promoter of HMGB1 in NcoR1-KD and NcoR1-OE cells. Cells were subjected to ChIP assays with anti-NcoR1, anti-SP1, or IgG. The purified DNA samples were subjected to RT-qPCR analyses to detect the occupancies of NcoR1 and SP1 on the promoter of HMGB1. * P < 0.05 and ** P < 0.01. (D) The relative occupancies of NcoR1 and SP1 on the promoter of HMGB1 in SP1-KD and SP1-OE cells. Cells were subjected to ChIP assays with anti-NcoR1, anti-SP1, or IgG. The purified DNA samples were subjected to RT-qPCR analyses to detect the occupancies of NcoR1 and SP1 on the promoter of HMGB1. * P < 0.05, ** P < 0.01 and *** P < 0.001.