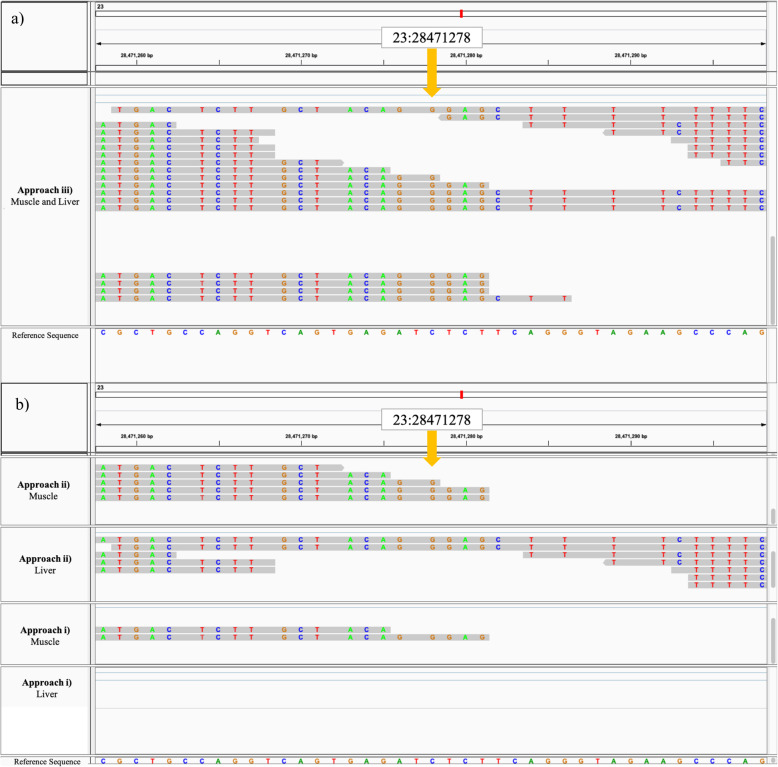
Fig. 2.
Visualization of the detection of an example variant (23: 28471278) using Approach iii), which is not detected by Approach i) or Approach ii), and corresponding read mapping. a Read mapping at example detected variant using Approach iii) Merged by RFI and tissue group; b Read mapping at example detected variant using Approach i) non-merged, and Approach ii) Merged by RFI group. Approach iii) Muscle and Liver: muscle and liver samples merged for low RFI .bam file. Approach ii) Muscle: merged muscle samples for low-RFI .bam file Approach ii) Liver: merged liver samples for low-RFI .bam file. Approach i) Muscle – non-merged individual muscle sample .bam file (sample accession number: ERS1342445). Approach i) Liver – non-merged individual liver sample .bam file (sample accession number: ERS579394). Approach descriptions: i) non-merged samples; ii) merged samples for low-RFI and merged samples for high-RFI for each tissue; iii) merged samples for low- and high-RFI for both tissues. Legend: Top numerical row (bp) = base pair position along transcriptome; bottom coloured row (bp letter) = UMD3.1 bovine reference genome (release 94) sequence. Coloured letters: Grey space = nucleotide base matches the reference base, Green = nucleotide base A, Red = nucleotide base T, Blue = nucleotide base C, Orange = nucleotide base G. Sequence region: Exon. Yellow arrow: Example variant at 23:28471278 detected by Approach iii) and not detected by Approach i) or Approach ii). Total read count coverage at variant site: Approach iii) = 10 (alternative allele = G (10), reference allele = C (0)); Approach ii) Muscle = 3 (alternative allele = G (3), reference allele = C (0)); Approach ii) Liver = 2 (alternative allele = G (2), reference allele = C (0)); Approach i) Muscle = 1 (alternative allele = G (1), reference allele = C (0)); Approach i) Liver = 0 (alternative allele = G (0), reference allele = C (0))