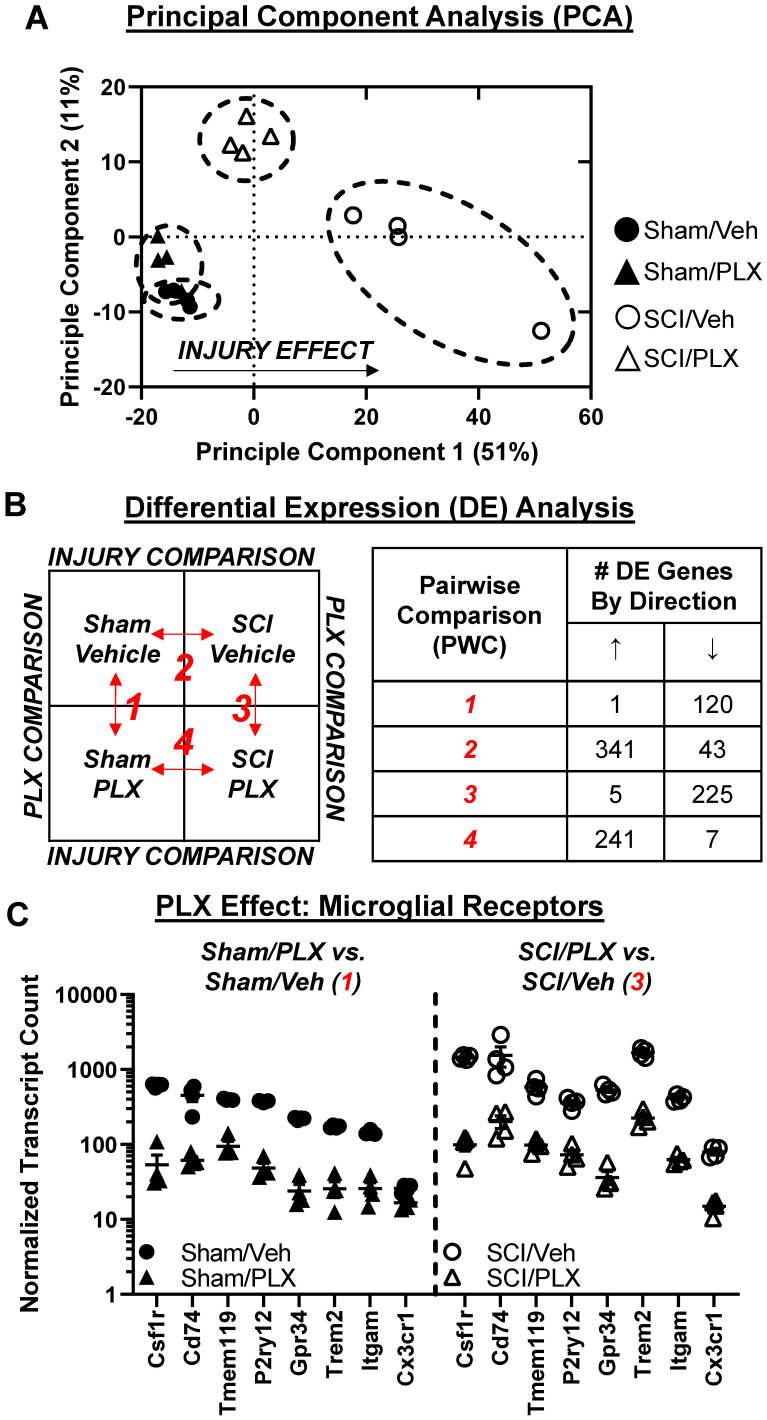
Figure 6.
Long-term depletion of microglia with PLX after SCI significantly alters the transcriptome at the injury site. A NanoString nCounter® Neuroinflammation panel was used to assess transcriptional changes at the spinal cord injury site at 6 weeks post-injury. (A) Principle component analysis (PCA) was performed using all normalized gene counts from the NanoString panel. The four sample groups were Sham/Veh (closed circles), Sham/PLX (closed triangles), SCI/Veh (open circles), and SCI/PLX (open triangles). PCA revealed distinct clustering (dashed ellipses) of the four sample groups across the first two principle components, PC1 and PC2, which accounted for 51% and 11%, respectively, of the total variation across samples. Injury-related effects were captured on PC1, separating the SCI/Veh group on the right from the Sham groups on the left. The SCI/PLX group was near the midline of the axis, indicating a reduction of the injury effect. (B) Differential expression (DE) analysis was performed on pairwise group comparisons using NanoStringDiff with a Benjamini-Hochberg false discovery rate correction (adjusted p-value < 0.05). Four pairwise comparisons were performed: (1) Sham/PLX vs. Sham/Veh; (2) SCI/Veh vs. Sham/Veh; (3) SCI/PLX vs. SCI/Veh; and (4) SCI/PLX vs. Sham/PLX. DE expressed genes in Injury Comparisons 2 and 4 had predominantly increased fold-change while PLX Comparisons 1 and 3 had predominantly decreased fold-change. (C) Plot of transcript counts for well-established microglial receptor markers (Csf1r, Cd74, Tmem119, P2ry12, Gpr34, Trem2, Itgam, Cx3cr1), indicating an effective depletion of microglia by PLX. All receptors had decreased gene expression in both comparisons (adjusted p-value < 0.05), except for Cx3cr1 in PLX Comparison 1 due to low baseline counts. n = 4 mice/group. Data presented as individual data points with mean ± S.E.M. Sham/Veh (closed circles), Sham/PLX (closed triangles), SCI/Veh (open circles), and SCI/PLX (open triangles).