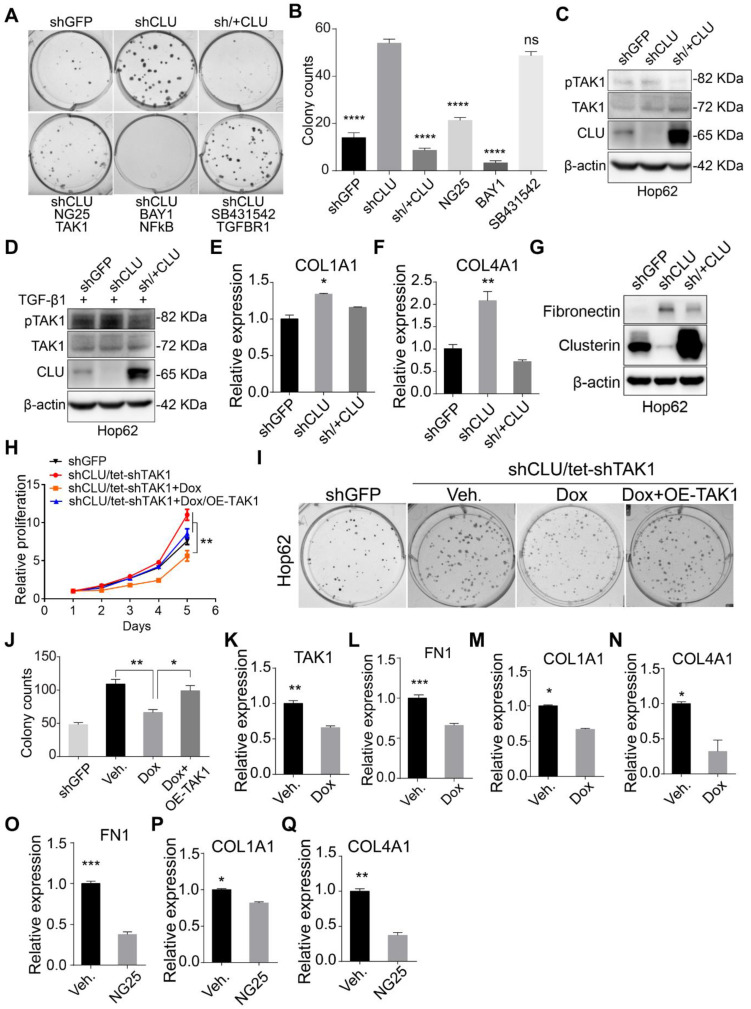
Figure 2.
CLU knockdown enhances TAK1 signaling. (A) TAK1 or NF-κB inhibitor specifically reduced colony forming ability of CLU knockdown lung cancer cell. 200 Hop62-shCLU cells were seeded and treated with indicated inhibitors for 7 days; NG25 (2.5 μM), BAY001 (1 μM), SB431542 (2.5 μM). Cell colonies were fixed and stained with 0.5% crystal violet in methyl alcohol. shGFP for control knockdown; shCLU for CLU knockdown; sh/+CLU for CLU re-expression in CLU knockdown cells. (B) Quantification of colony numbers. Statistic with one-way ANOVA test. (C) and (D) Impact of CLU expression on TAK1 activity. Hop62 cells were treated (C) without or (D) with TGF-β1 (5 ng/mL) for 2 h. Phosphor-TAK1, total TAK1 and CLU protein level was detected with indicated antibodies. (E) and (F) RT-PCR analysis of impact of CLU knockdown or re-expression on COL1A1 and COL4A1 expression in Hop62 cells. Total RNA of Hop62 cells with indicated were extracted and COL1A1 and COL4A1 expression was detected with RT-PCR. (G) Impact of CLU expression on Fibronectin expression in lung cancer cells. Fibronectin and CLU levels were assayed through Western analysis. (H) Impact of TAK1 knockdown or re-expression on cell proliferation of CLU knockdown Hop62 (Hop62-shCLU/tet-shTAK1) cells. TAK1 knockdown in Hop62-shCLU cells was induced with 1 μg/mL Dox. Statistic shown on day 5 with two-tailed t-test. (I) Impact of TAK1 knockdown on 2-D colony formation of CLU knockdown Hop62 cells. Cell colonies were fixed and stained with 0.5% crystal violet in methyl alcohol. (J) Quantification of I, one-way ANOVA test. (K-N) RT-PCR analysis of impact of TAK1 knockdown on FN1, COL1A1 and COL4A1 expression in CLU knockdown Hop62 cells. Hop62-shCLU cells were treated with 1 μg/mL Dox for TAK1 inducible knockdown. Statistic with two-tailed t-test. (O-Q) RT-PCR analysis of the impact of TAK1 inhibition with NG25 on FN1, COL1A1 and COL4A1 expression. Hop62-shCLU cells were treated with NG25 (2.5 μM) for 24 h before detecting mRNA level. *P < 0.05, **P < 0.005, ***P < 0.0001; Data plotted are mean ± s.e.m.; n = 3.