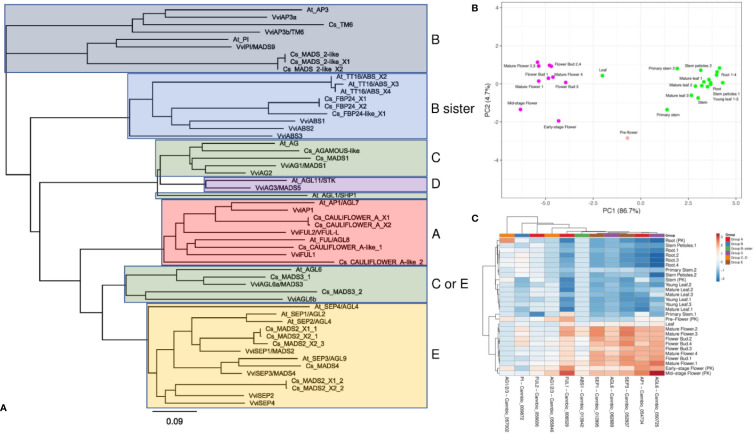
Figure 3.
(A) Similarity-based neighbor-joining analysis performed using 21 amino acid sequences from the C. sativa (Cs) proteome (GCA_900626175.1) selected for their putative orthology (Table 2 and, more specifically, Supplementary Table 2) with well-characterized ABCDE MADS box proteins belonging to Arabidopsis thaliana (At) and Vitis vinifera (Vvi). (B) Taking advantage of a recent in silico analysis of 31 RNA-seq datasets derived from different tissues of two different psychoactive strains (Finola and Purple Kush, NCBI SRA accession numbers: SRP006678 and SRP008673) of C. sativa (Massimino, 2017), a principal component analysis was performed using the expression values of the MADS box genes previously identified. The analysis is based on the ln(x+1) transformed (RPKM) values (reads per kilobase of transcript per million mapped reads) and showed a clear separation of samples related to reproductive organs from those related to vegetative organs. (C) Heat map showing the relative expression of each gene in the different tissues considered.