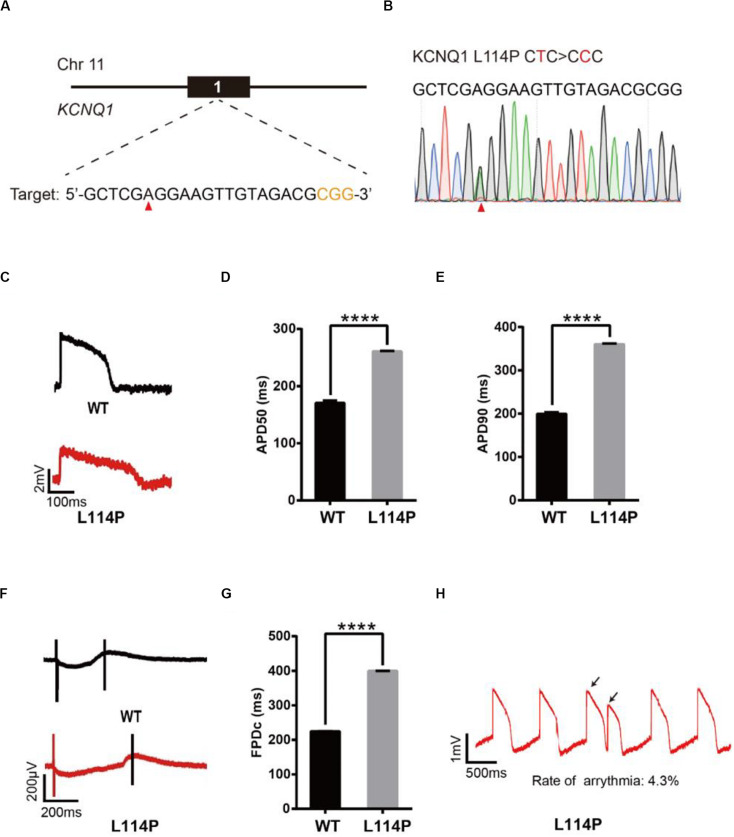
FIGURE 2.
Base editing of L114P-KCNQ1 for LQT1 modeling. (A) L114P target site on KCNQ1. “CGG” PAM sequence is shown in orange; target nucleotide to be edited is indicated by a red triangle. (B) A heterozygous clone is confirmed by Sanger sequencing. The mutated nucleotide is shown in red; the mutated nucleotide is indicated by a red triangle. (C) Single trace of action potentials in WT-CMs and L114P-CMs. (D,E) Quantification of action potential at APD50 and APD90. n = 3 independent experiments, unpaired t-test, P < 0.0001. (F) Signals of field potential duration recorded by MEA. (G) Quantification of corrected field potential durations (FPDc). n = 3 independent experiments, unpaired t-test, P < 0.0001. (H) Representative traces of action potentials. The abnormal AP signals were labeled by black arrows. A value of P < 0.05 was considered to be statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).