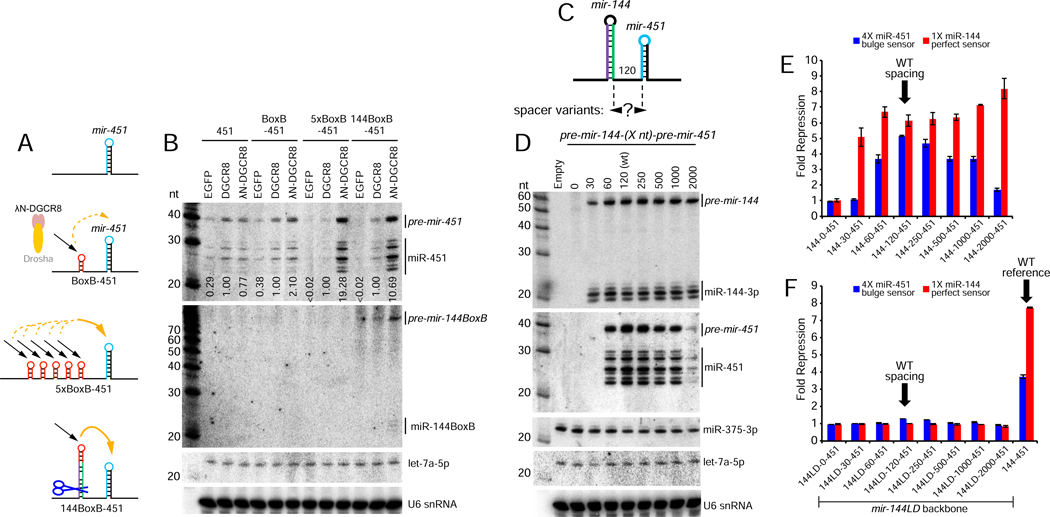
Figure 4. Local recruitment and transfer of Microprocessor promotes nuclear mir-451 biogenesis.
(A) Schematics of constructs tested. Using Δ144–451 as a starting construct, we inserted 1 or 5x BoxB elements at the site of the mir-144 hairpin. We also made a version in which BoxB was introduced at the terminal loop of mir-144 (144BoxB-451). (B) Northern blotting shows that λN-DGCR8 is slightly better than DGCR8 at promoting biogenesis of BoxB-451, but is far better on the 5xBoxB-451 substrate (almost 20-fold). Notably, the single BoxB in 144BoxB-451 yields much better enhancement of miR-451 biogenesis with λN-DGCR8 compared to DGCR8 (10 fold). (C) Varying the spacer length between mir-144 and mir-451. Note that these distances correspond to the nts between Drosha cleavages and therefore the “0” nt spacing actually deletes all the lower sequences between these miRNAs while the “30” nt spacing removes one side of the lower single-stranded flanking sequences for both miRNAs. (D) mir-144/451 and mir-375 expression constructs were cotransfected and blotted sequentially; endogenous let-7a and U6 snRNA were also assayed as further loading controls. The biogenesis of miR-144 is relatively stable across these length variants (excepting “0”, which is expected to be non-functional for both miRNAs), whereas biogenesis of miR-451 was optimal at its normal spacing and gradually declined with greater inter-miRNA distance. Little miR-451 was produced at a 2kb spacing. No miR-451 was produced at the 30-nt spacing, which would leave insufficient flanking single-stranded sequence for Microprocessor to recognize pri-mir-451 following mir-144 cropping. (E) Sensor assays of the mir-144-(Xnt)-451 length variants. The activity of miR-451 declines with increasing distance from mir-144. (F) Enhancement at a longer distance. Although tempered, mir-144 still enhances mir-451 biogenesis across a range of longer spacer distances. These are all due specifically to mir-144, since deletion of the mir-144 loop (144LD) within all of these constructs abolishes both miR-144 and miR-451 function.