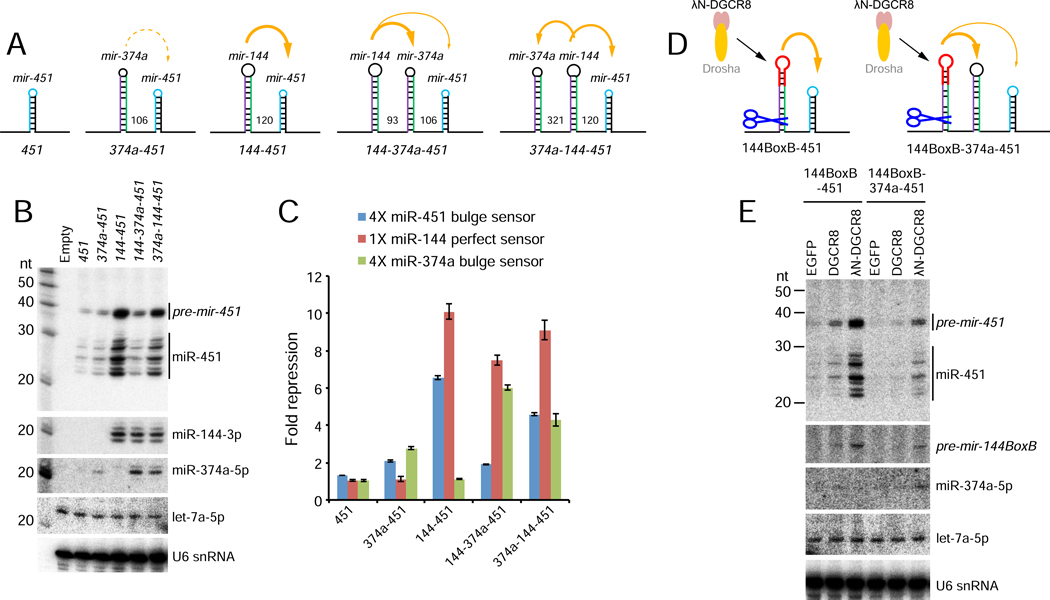
Figure 7. Location-dependent competition amongst suboptimal clustered miRNAs.
(A) Schematics of miRNA constructs. In addition to Δ144–451 solo and mir-144/451 constructs used earlier, we replaced mir-144 with suboptimal mir-374a (374a-451), inserted mir-374a in between mir-144 and 451 (144–374a-451), or placed it upstream in this operon (374a-144–451). (B) Northern blotting shows that mir-374a only modestly enhances mir-451 biogenesis. However, miR-374a can be enhanced by mir-144, but in so doing, it blocks biogenesis of downstream mir-451. Placing mir-144 in between these suboptimal miRNAs allows both to experience some biogenesis enhancement. (C) Sensor assays of these constructs correlate well with the Northern results, indicating location-dependent competition between mir-374a and mir-451 for benefitting from proximity to mir-144. Reciprocally, miR-144 activity is little affected by the presence or number of suboptimal neighbors. (D) In vivo test of competition for locally released Microprocessor by suboptimal primary miRNA hairpins. λN-DGCR8 is tethered to the terminal loop of mir-144 and its capacity to locally promote the biogenesis of neighboring suboptimal miRNAs is assayed; untethered DGCR8 is used as a control. (E) Northern blotting shows that local recruitment and release of λN-DGCR8 from neighboring pre-mir-144BoxB can strongly potentiate mir-451 biogenesis, but this is blocked by proximal location of suboptimal mir-374a.