Fig. 4 |. Capsid accommodation of the DNA-translocating portal complex and periportal CATCs.
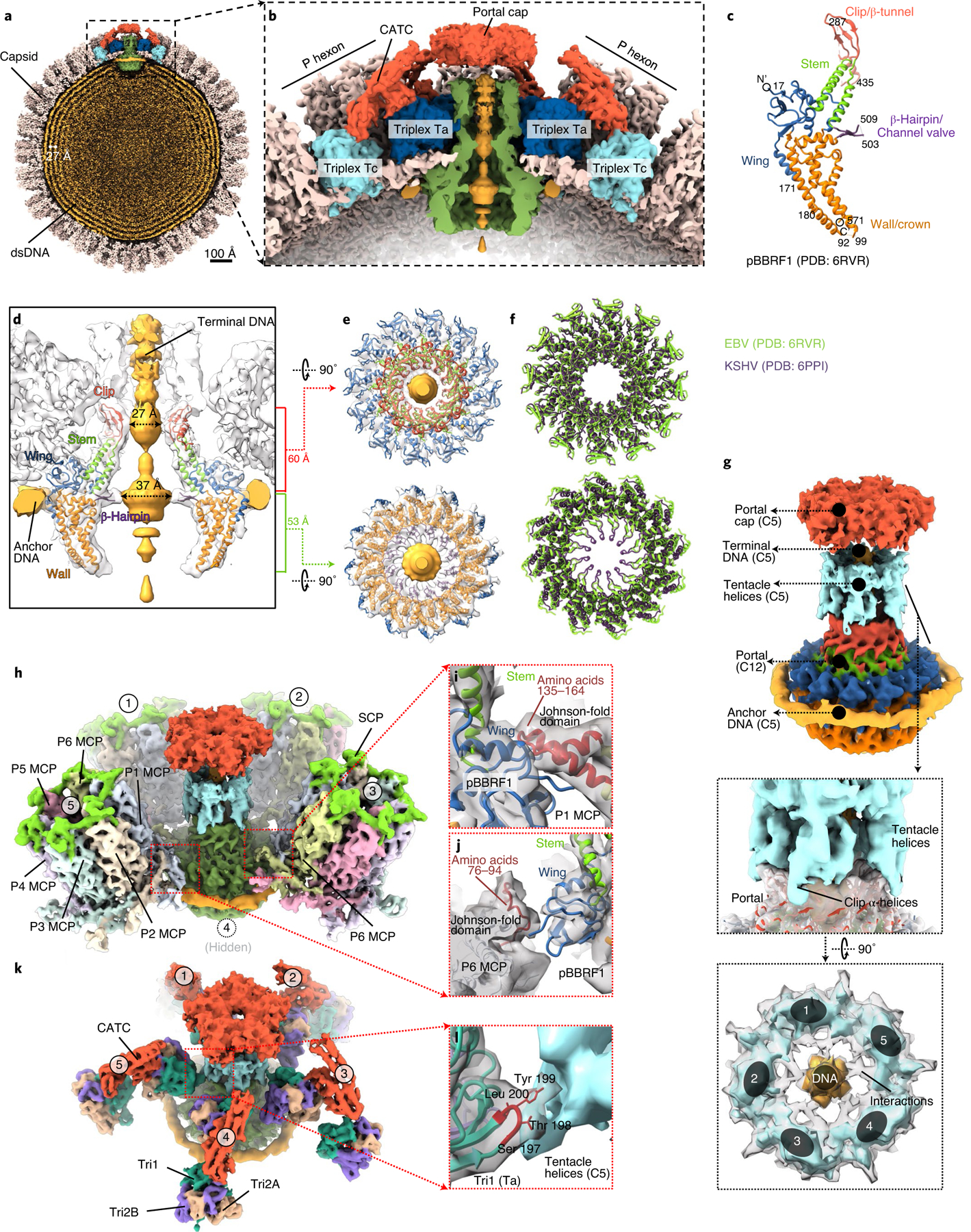
a,b, Clipped (a) and zoomed-in (b) views of the C5 whole-virus reconstruction, showing packaged dsDNA within the capsid with neighbouring dsDNA duplexes spaced ~27 Å apart (a) and structural components around the portal vertex (b). c, Atomic model of the recombinant portal protein9 shown as a monomer coloured by domains. d–f, Clipped view of the portal vertex region showing the fitted atomic models (ribbon) of two opposing subunits (d) and the two constrictions along the DNA-translocating channel (e). The superposition (f) of eBV and KSHV portal atomic models reveals similarities along these constrictions. g–l, Composite map of eBV portal region, showing interactions of the portal complex and DNA, and the MCP and Tri1. The C12 portal subparticle reconstruction was placed into the C5 portal vertex subparticle reconstruction by referencing HSV-1 and KSHV C1 portal vertex structures18,19, showing DNA, tentacle helices and portal cap structures surrounding the fitted atomic model of the portal complex (g). Five surrounding P hexons (h) interact with the wing domain of the portal protein through amino-acid segments 135–164 (i) and 76–94 (j) of P1 MCP and P6 MCP, respectively. Both segments are located within the Johnson-fold domain of the MCP. Likewise, surrounding the structure shown in g are five Ta triplexes (k), the Tri1 subunit of which interacts at residues 198 and 199 with the tentacle helices (l).
