Abstract
Carbapenemase-producing Klebsiella pneumoniae emerged as a nosocomial pathogen causing morbidity and mortality in patients. For infection prevention it is important to track the spread of K. pneumoniae and its plasmids between patients. Therefore, the major aim was to recapitulate the contents and diversity of the plasmids of genetically related K. pneumoniae strains harboring the beta-lactamase gene blaKPC-2 or blaKPC-3 to determine their dissemination in the Netherlands and the former Dutch Caribbean islands from 2014 to 2019. Next-generation sequencing was combined with long-read third-generation sequencing to reconstruct 22 plasmids. wgMLST revealed five genetic clusters comprised of K. pneumoniae blaKPC-2 isolates and four clusters consisted of blaKPC-3 isolates. KpnCluster-019 blaKPC-2 isolates were found both in the Netherlands and the Caribbean islands, while blaKPC-3 cluster isolates only in the Netherlands. Each K. pneumoniae blaKPC-2 or blaKPC-3 cluster was characterized by a distinct resistome and plasmidome. However, the large and medium plasmids contained a variety of antibiotic resistance genes, conjugation machinery, cation transport systems, transposons, toxin/antitoxins, insertion sequences and prophage-related elements. The small plasmids carried genes implicated in virulence. Thus, implementing long-read plasmid sequencing analysis for K. pneumoniae surveillance provided important insights in the transmission of a KpnCluster-019 blaKPC-2 strain between the Netherlands and the Caribbean.
Subject terms: Computational biology and bioinformatics, Microbiology
Introduction
Antimicrobial resistance is spreading rapidly among Enterobacterales, including Klebsiella pneumoniae, Escherichia coli and Enterobacter spp.1. Within the cell, extra-chromosomal DNA such as plasmids encode genes that confer resistance to last resort antibiotics, including carbapenems and colistin, and can transfer between Enterobacterales2. Currently, carbapenemase-producing Enterobacterales (CPE) rank among the most problematic nosocomial pathogens with limited outlook on novel effective therapeutics3,4. With the current increase of multidrug-resistant infections with CPE worldwide, total healthcare costs are anticipated to increase. K. pneumoniae is often referred to as the “canary in the coalmine”, as new antimicrobial resistance (AMR) genes have been associated with K. pneumoniae in the first clinical reports prior dispersal of the AMR genes among other Gram-negative bacteria5. Most newly acquired AMR genes of K. pneumoniae are the result of horizontal gene transfer through conjugative plasmids6–8. The K. pneumoniae carbapenemase KPC encoded by the blaKPC gene is an Ambler class A serine carbapenemase, which is often located on a transmissible plasmid-associated transposon Tn4401, or variants hereof9–12. Tn4401 consists of flanking imperfect repeat sequences, a Tn3 transposase gene, a Tn3 resolvase gene and the ISKpn6 and ISKpn7 insertion sequences10. The blaKPC-2 and blaKPC-3 carbapenemases are the most commonly identified variants that have spread globally and provide resistance to penicillins, carbapenems, cephalosporins, cephamycins and monobactams13,14. The KPC-2 and KPC-3 carbapenemases differ in only one amino acid as a histidine at position 272 is mutated to tyrosine (H272Y) in the KPC-3 variant15.
CPE isolates including K. pneumoniae are routinely send to the National Institute for Public Health and the Environment (RIVM) and are typed by Illumina next-generation sequencing (NGS) in the Dutch National CPE Surveillance program to identify AMR genes and to determine possible transmission of strains16. NGS typically yields short sequence reads of 150 bases, thereby hampering the assembly of complete chromosomes and plasmids17. This is often due to large mobile genetic elements, such as insertion sequence elements, transposons, and other repetitive sequences e.g. tandem repeat regions of > 1500 bp in size. However, combining Illumina NGS sequencing with long-read third generation sequencing (TGS), which produces 1000 to 500,000 bases or longer sequence reads, can overcome this problem and enables the reconstruction of chromosomes and complete plasmids18,19. Currently, the transmission of K. pneumoniae between persons in the Netherlands and the Caribbean and the impact hereof is not thoroughly understood. Five percent of the K. pneumoniae isolates collected in the Dutch National CPE Surveillance Program are retrieved from the Caribbean. It is also not clear whether plasmids of K. pneumoniae circulate endemically in the Netherlands or are introduced from the Caribbean. blaKPC-type K. pneumoniae represent the third largest group (17.5%) of the K. pneumoniae isolates collected in the Dutch National CPE Surveillance Program after the blaOXA-48-type (48.5%) and blaNDM-1-type (24.3%) K. pneumoniae. While the prevalence of carbapenemase-producing K. pneumoniae and associated infections in the Netherlands is relatively low, the establishment of genomic surveillance of K. pneumoniae using TGS is of high importance20,21. It provides for insights in the transmission of specific strains containing plasmids with AMR genes and/or virulence determinants. We therefore investigated the distribution of K. pneumoniae cluster isolates harboring blaKPC-2 or blaKPC-3 alleles obtained from the Dutch National CPE Surveillance Program and analyzed the contents of its plasmids using long-read third-generation sequencing.
Results
Distribution and genetic relationship of blaKPC-2 and blaKPC-3 carrying K. pneumoniae
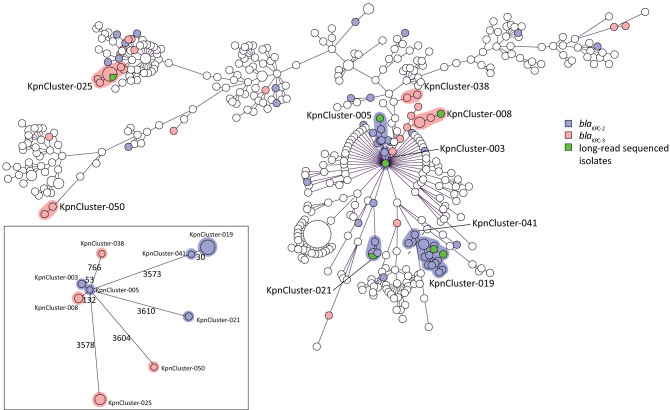
A collection of 478 carbapenemase-producing K. pneumoniae isolates submitted to the Dutch National CPE Surveillance program from January 1st 2014 until June 30th 2019 to the National Institute for Public Health and the Environment (RIVM) were included in this study. The study collection comprised 84 K. pneumoniae blaKPC-positive isolates of which 51 contained the blaKPC-2 allele and 33 harbored the blaKPC-3 allele (Table 1). Sixty isolates originated from the Netherlands and 24 isolates originated from the Caribbean. Of the 24 Caribbean isolates, 22 carried the blaKPC-2 allele and only two contained the blaKPC-3 allele. Whole genome multi-locus sequence typing (wgMLST), using an in-house wgMLST scheme based on 4,978 genes, of the 478 carbapenemase-producing K. pneumoniae isolates collected in the RIVM revealed that 23 K. pneumoniae blaKPC-2 isolates grouped together in five distinct genetic clusters. Fifteen K. pneumoniae blaKPC-3 isolates grouped in four distinct clusters which were obtained from the Netherlands and 46 isolates were unrelated. The K. pneumoniae cluster isolates (termed KpnClusters) had unique classical MLST sequence types, of which ST144 (KpnCluster-021) and ST560 (KpnCluster-019) were not described previously (Table 1, Fig. 1). KpnCluster-003 and KpnCluster-005 were comprised of five K. pneumoniae blaKPC-2 isolates that were exclusively obtained from the Netherlands, while KpnCluster-021 and KpnCluster-041 contained five isolates from the Caribbean. The majority (n = 10) of the KpnCluster-019 isolates were obtained from the Caribbean. However, three isolates were from the collection of the Netherlands. One person from whom a KpnCluster-019 isolate was retrieved in August 2017 in the Netherlands, lived in the Caribbean until June 2017 and migrated to the Netherlands in July, demonstrating intercontinental transmission. No epidemiological data could be retrieved from the other two Dutch KpnCluster-019 isolates. Furthermore, most genetic clusters were only distantly related to each other (Fig. 1). Based on wgMLST, the genetic distance between KpnCluster-019 and KpnCluster-041 was 30 alleles (0.6%) and for KpnCluster-003 and KpnCluster-005 53 alleles (1.1%). KpnCluster-008 differed 132 alleles (2.65%) from KpnCluster-005. While the allelic difference between these clusters was low, the other genetic clusters differed 3573 to 3610 alleles (71.8–72.5%) from KpnCluster-005. This confirmed that most clusters were unrelated, and it is in line with the location of these genetic clusters in the minimum spanning tree.
Table 1.
Distribution of K. pneumoniae blaKPC-2 and blaKPC-3 isolates and resistance to meropenem.
| blaKPC allele | KpnCluster | MLST | The Netherlands | Caribbean | Total | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence type | S | I | R | S | I | R | |||
| blaKPC-2 | KpnCluster-003 | ST258 | 2 | 2 | |||||
| KpnCluster-005 | ST258 | 3 | 3 | ||||||
| KpnCluster-019 | ST560 | 1 | 2 | 6 | 4 | 13 | |||
| KpnCluster-021 | ST144 | 1 | 2 | 3 | |||||
| KpnCluster-041 | ST560 | 2 | 2 | ||||||
| Non-KpnCluster | variant | 3 | 4 | 14 | 1 | 2 | 4 | 28 | |
| Subtotal | 51 | ||||||||
| blaKPC-3 | KpnCluster-008 | ST512 | 4 | 4 | |||||
| KpnCluster-025 | ST307 | 1 | 6 | 7 | |||||
| KpnCluster-038 | ST11 | 2 | 2 | ||||||
| KpnCluster-050 | ST13 | 1 | 1 | 2 | |||||
| Non-KpnCluster | variant | 3 | 5 | 8 | 1 | 1 | 18 | ||
| Subtotal | 33 | ||||||||
| Total | 8 | 12 | 40 | 10 | 7 | 7 | 84 | ||
Based on the clinical breakpoints according to EUCAST, the isolates were classified as sensitive (S; < 2 mg/L), intermediate (I; ≥ 2 to 8 mg/L) and resistant (R; > 8 mg/L).
Figure 1.
Minimum spanning tree based on wgMLST of 478 sequenced K. pneumoniae isolates. Circles represent K. pneumoniae isolates, and the sizes of the circles indicate the numbers of isolates. Lines connecting the circles represent the genetic distance in numbers of alleles; the longer the connecting line, the larger the genetic distance. K. pneumoniae blaKPC-2 isolates were marked blue and K. pneumoniae blaKPC-3 were marked magenta. K. pneumoniae blaKPC-2 or blaKPC-3 cluster isolates that were sequenced with TGS were marked green. Genetic clusters were indicated with either a blue or a magenta halo around the circles, if two or more isolates differ ≤ 20 alleles. A categorical coefficient was used for the clustering. Cluster names are indicated. Inset: genetic distance between the KpnClusters in which the allelic difference is indicated by numbers.
The resistome diversity among genetic clusters
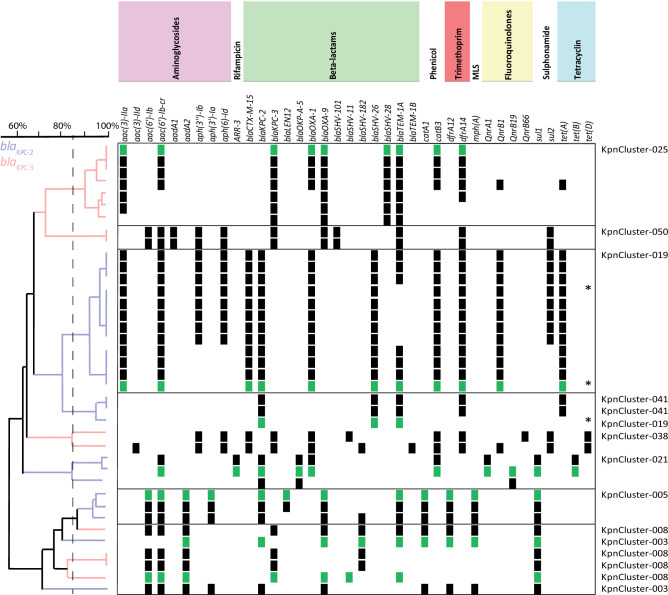
Analysis of the NGS-derived resistomes of the cluster and non-cluster isolates showed that K. pneumoniae harbored either the blaKPC-2 or the blaKPC-3 allele, none of the isolates carried both alleles (Fig. 2, Suppl. Figure 1). All of the K. pneumoniae isolates contained the fosA, oqxA and oqxB genes. An unweighted hierarchical clustering (UPGMA) based on the presence or absence of AMR genes revealed that most genetic cluster isolates group together per cluster, since the resistomes were more than 85% similar. In contrast to this, the resistomes of the non-cluster isolates were very diverse and less related since the resistomes of these isolates were less than 85% similar (Suppl. Figure 1). Likewise, the resistomes of one group of K. pneumoniae KpnCluster-003 blaKPC-2 and KpnCluster-008 blaKPC-3 cluster isolates with 53 to 132 alleles difference were also unrelated. KpnCluster-019 isolates are unique when compared to the blaKPC-2 clusters KpnCluster-003, KpnCluster-005, and KpnCluster-021, in that they carried aminoglycoside (aac(3)-IIa), extended spectrum beta-lactams (blaCTX-M-15, blaSHV-26), fluoroquinolone (qnrB1) and tetracyclin (tetA) antimicrobial resistance (AMR) genes. KpnCluster-019 and KpnCluster-041 isolates, obtained from the Caribbean, were closely related based on wgMLST, and group together based on the resistome too. The absence of AMR genes aph(3′')-Ib, aph(6)-Id and sul2 in five of KpnCluster-019 isolates, including the TGS sequenced isolates, indicate the absence of an AMR gene containing plasmid. In addition, the presence of three KpnCluster-019 isolates from the Netherlands with varying resistomes within the cluster suggests additional transmissions. KpnCluster-025 blaKPC-3 isolates contained the aminoglycoside (aac(3)-IIa) and beta-lactam AMR genes (blaSHV-28), while the other Kpn blaKPC-3 clusters did not. Notably, mcr genes conferring resistance to colistin were not detected in the 84 isolates analyzed. The majority of the K. pneumoniae blaKPC-2 and blaKPC-3 isolates were resistant to meropenem (47/84; 56%). More specifically, seven of the 23 K. pneumoniae blaKPC-2 cluster isolates (30%) and 13 of the 15 blaKPC-3 cluster isolates (87%) were resistant to meropenem. The remainders of the cluster and non-cluster isolates were intermediate resistant or sensitive for meropenem (Table 1).
Figure 2.
Resistome of K. pneumoniae blaKPC-2 and blaKPC-3 cluster isolates. K. pneumoniae blaKPC-2 and blaKPC-3 cluster isolates were indicated on the y-axis and AMR genes on the x-axis. All isolates analysed contained the fosA, oqxA and oqxB AMR genes and were not included in this figure. The clustering was based on the presence (squares) and absence of AMR genes. Antibiotic classes are indicated above the AMR genes in different colors. Resistance genes in K. pneumoniae blaKPC-2 or blaKPC-3 cluster isolates that were sequenced with TGS were marked with green squares. Genetic relatedness was depicted in an UPGMA tree in which K. pneumoniae blaKPC-2 isolates were marked with blue branches, and K. pneumoniae blaKPC-3 were marked magenta. Dutch KpnCluster-019 isolates were marked with an *. A dotted line marks the 85% cut off.
Antibiotic resistance genes among the genomic elements of the distinct genetic clusters
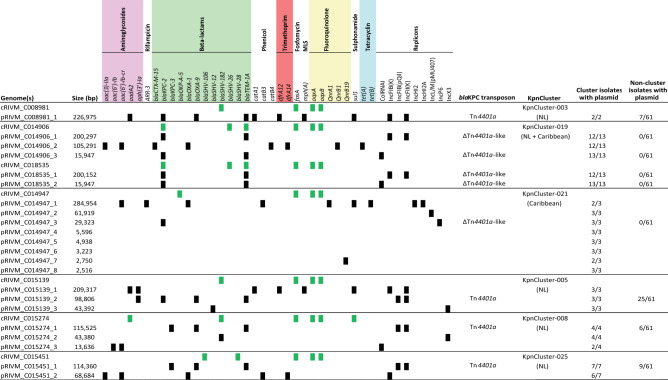
Long-read sequencing of seven isolates from six of the nine genetic K. pneumoniae blaKPC clusters with ≥ 3 isolates per cluster, revealed 22 plasmids with varying sizes (Fig. 3). Plasmids containing either the blaKPC-2 or blaKPC-3 allele were diverse in size. The large (≥ 150–250 kb) and medium (≥ 50–150 kb) sized plasmids contained one or two replicons from the incompatibility group IncFIB(K) and IncFII(K), IncHI2 and IncHI2a, or IncFIB(pQil) (Fig. 3). The small plasmids (< 50 kb) contained ColRNAI or IncX3/IncL/IncP6 type of replicons. The chromosomes of the analyzed isolates contained on average five acquired AMR genes, while the plasmids contained on average nine AMR genes. Sixteen of the 22 plasmids contained AMR genes from various classes and five plasmids from the isolate of KpnCluster-021 did not. The AMR genes conferring resistance to phenicol, trimethoprim and macrolide antibiotics were located only on medium or large sized plasmids. The small plasmids had one or two AMR genes conferring resistance to aminoglycosides or beta-lactams. Resistance genes for fosfomycin (fosA) and fluoroquinolones (oqxA and oqxB) were exclusively located on the chromosomes of the seven cluster isolates. KpnCluster-019 and KpnCluster-021 associated with the Caribbean contained plasmids encoding genes for phenicol and tetracyclin resistance. The KpnCluster-019 and KpnCluster-021 plasmids were not found in non-cluster isolates, whereas the plasmids of the other clusters were detected in a subset non-cluster isolates (Fig. 3). The plasmids of KpnCluster-003 and KpnCluster-005 were present in each of its cluster isolates, however, in isolates of the other clusters occasionally plasmids were lost, thereby impacting the composition of the resistome (Figs. 2 and 3).
Figure 3.
Antimicrobial resistance genes on chromosomes and plasmids. The presence of AMR genes among the 22 plasmids of seven TGS sequenced isolates is indicated with black squares and for the chromosomes using green squares. Chromosomes (cRIVM_C0xxxx) and plasmids (pRIVM_C0xxxx) are depicted on the Y-axis, and AMR genes on the x-axis. Antibiotic classes are indicated above the AMR genes in different colors.
The blaKPC-2 KpnCluster-019 isolates were obtained from both the Caribbean and the Netherlands, while blaKPC-2 KpnCluster-021 isolates originated only from the Caribbean (Table 1, Fig. 3). In the KpnCluster-019 isolate RIVM_C014906, three copies of the blaKPC-2 gene were present, while other cluster isolates had only one blaKPC copy. One copy was located in the chromosome, one copy in the 200 kb plasmid pRIVM_C014906_1 and a third copy on the 16 kb plasmid pRIVM_C014906_3. All these three blaKPC-2 copies were located on a highly similar Tn4401a-derived ∆Tn4401a-like transposon of 5.6 kb in this strain. The chromosomes contained this ∆Tn4401a-like transposon in the exact same region. KpnCluster-003, KpnCluster-005, KpnCluster-008 and KpnCluster-025 consist of isolates that were obtained in the Netherlands and in these isolates the blaKPC allele was located on a Tn4401a transposon of 10 kb.
Comparison of the K. pneumoniae plasmid content among clusters
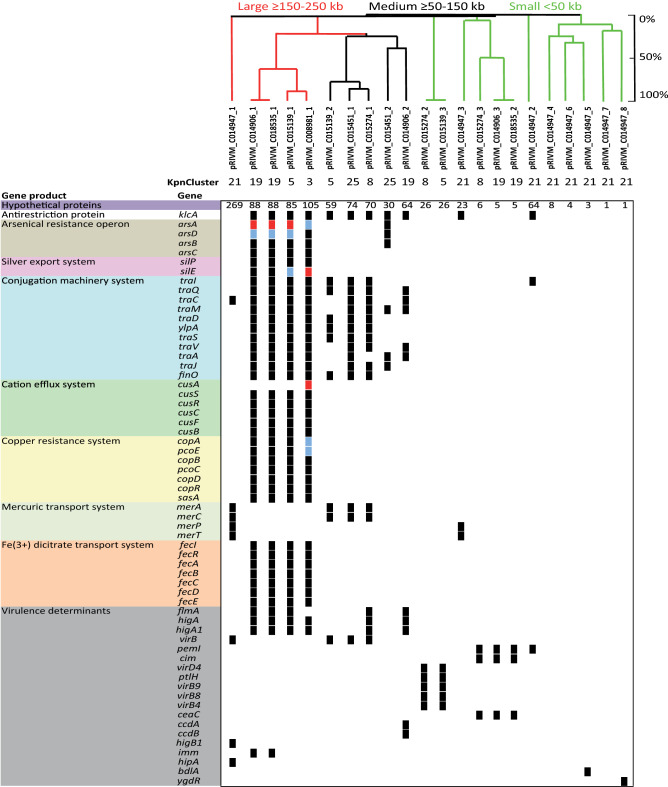
An UPGMA clustering based on the DNA sequence of the 22 plasmids from distinct genetic clusters revealed that the majority of the plasmids were unrelated (Fig. 4). The largest two plasmids pRIVM_C008981_1 from KpnCluster-003 and pRIVM_C014947_1 from Kpncluster-021 carried the largest number of genes and this number decreased by the decreasing size of the plasmids. Most these plasmid located genes had unknown function. The large and medium sized plasmids contained the klcA gene, encoding an antirestriction protein implicated in the facilitation of blaKPC allele transfer22. None of the plasmids contained known virulence determinants such as rmpA, rmpA2, iroBC, or iucABC implicated in hypervirulence23,24. Comparison of the large plasmids revealed that pRIVM_C008981_1 and pRIVM_C015139_1 from KpnCluster-003 and KpnCluster-005 displayed 90% similarity (Fig. 4). Plasmid pRIVM_C014947_1 from KpnCluster-021 was not related to any other of the large plasmids. Despite the low similarity, these large plasmids from KpnClusters-003, -005, and -019, shared important clusters of genes among them. They all contained the silE and silP genes encoding a silver-binding protein and a silver exporting ATPase, cusSRCFB genes implicated in cation efflux, the copABCD-pcoE genes involved in copper resistance and the arsHACBAD arsenic resistance gene cluster. These large plasmids also contained fecIRABCDE implicated in Fe(3+)-dicitrate transport, the traIDSQCVAJM-ylpA plasmid conjugation gene cluster, and the higA-higA1 antitoxins, except pRIVM_C014947_1 from KpnCluster-021. In addition, the large plasmids also contained a proportion of plasmid-specific and thus K. pneumoniae cluster specific gene content (Suppl. Figure 2).
Figure 4.
K. pneumoniae plasmid gene content. An UPGMA clustering was performed based on the plasmid DNA sequence for the determination of the genetic relation among the 22 plasmids. Similarity is indicated on the y-axis using a scale from 0 (not similar) to 100% (identical). A similarity of ≥ 85 to 100% is regarded as the same plasmid. The plasmids are indicated on the x-axis. The presence (black squares) and absence is indicated of annotated genes among the 22 plasmids of seven TGS sequenced isolates. If a gene was present twice, blue squares were used and more than 2, red squares were used. Colors indicated different groups of genes with a specific function. In the UPGMA tree, large plasmids are indicated in red, medium plasmids in black and small plasmids in green color.
Three medium-sized plasmids contained the virB virulence regulon transcriptional activator and the merAC mercuric reductase and transport protein. While pRIVM_C015274_1 from KpnCluster-008 and pRIVM_C015451_1 from KpnCluster-025 contained a plasmid conjugation gene cluster, plasmids pRIVM_C014906_2, pRIVM_C015139_2, and pRIVM_C015451_2 contained truncated versions hereof. The more distantly related pRIVM_C014906_2 plasmid from KpnCluster-019 had in addition to the higA-higA1 antitoxins also a ccdA-ccdB toxin-antitoxin system. The small plasmids (< 50 kb) contained genes implicated in virulence. Plasmids pRIVM_C015139_3 and pRIVM_C015274_2 displayed 99% similarity and carried the virD4-B9-B8-B4-ptlH Type IV secretion system. pRIVM_C014947_3 contained a merPT mercuric transport system, while pRIVM_C014906_3 and pRIVM_C015274_3 carried a ceaC colicin-E3. The plasmid pRIVM_C014947_5 contained the bdlA gene encoding a biofilm dispersion protein.
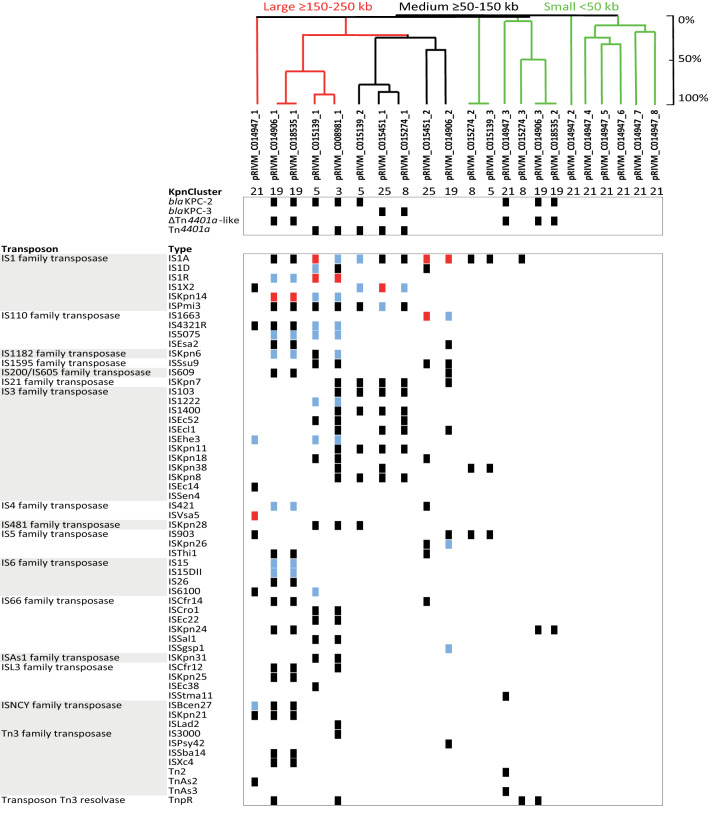
Transposable elements in K. pneumoniae plasmids from distinct clusters
The large and medium sized plasmids contained the most transposase sequences, and each plasmid had its unique transposon signature (Fig. 5). The IS1, IS110 and IS3 transposase families dominated in the K. pneumoniae plasmids among genetic clusters. The IS1 family transposase was found most frequently among the plasmids and in most copies within plasmids. In the large and medium sized plasmids, the blaKPC allele was located on a Tn4401a transposon, except in pRIVM_C014906_1 and pRIVM_C01835_1 from KpnCluster-019. In the small plasmids carrying a blaKPC, the carbapenemase allele was located on a ∆Tn4401a-like transposon. The large plasmids pRIVM_C008981_1, pRIVM_C015139_1 and pRIVM_C014906_1 from KpnClusters-003, -005, and -019 harbored 37, 32 and 31 annotated tranposases, respectively. In contrast, the largest plasmid pRIVM_C014947_1 of 285 kb from KpnCluster-021 contained only 16 transposons. The remainder of the plasmids from KpnCluster-021 also contained very few transposase sequences, in contrast to the other plasmids from the different clusters. The highly related pRIVM_C015139_3 and pRIVM_C015274_2 plasmids (99% similarity) from KpnCluster-005 and KpnCluster-008 had identical transposons. While IS66 and IS110 family transposase sequences also dominate in the large plasmids, the medium sized plasmids contained IS3 family type of transposases. The medium sized plasmids contained eleven to 23 transposases, and the small plasmids less than ten.
Figure 5.
K. pneumoniae plasmid-localized transposases. The presence (black squares) and absence is indicated of annotated transposases among the 22 plasmids of six TGS sequenced isolates. The plasmids are indicated on the x-axis. If a transposon was present twice, blue squares were used and more than 2, red squares were used. The light grey area indicates specific transposons found in only one plasmid. In the UPGMA tree, large plasmids are indicated in red, medium plasmids in black and small plasmids in green color.
Similarity with previously reported plasmids
BLAST analysis of the K. pneumoniae plasmids identified in this study showed that 15 of the 22 plasmids were similar to previously reported plasmids in the NCBI sequence database (Table 2). These plasmids covered five distinct genetic clusters, except pRIVM_C008981_1 from KpnCluster-003. To date, none of these plasmids of K. pneumoniae were reported to be implicated in healthcare-associated outbreaks. Plasmids pRIVM_C008981_1, pRIVM_C014906_1, pRIVM_C014906_3 containing blaKPC-2 and pRIVM_C015274_1 harboring blaKPC-3 from distinct genetic clusters only had low sequence coverage 35–87% with plasmids present in the NCBI sequence database. The other blaKPC-2 and blaKPC-3 plasmids had high (93–99%) sequence coverage, indicating that these similar plasmids were detected previously by other researchers. Plasmids pRIVM_C014906_2, pRIVM_C015139_1, pRIVM_C015274_2 and pRIVM_C015274_3, not carrying a blaKPC allele, displayed 97–100% sequence coverage and 99–100% identity to plasmids isolated from K. pneumoniae from different countries (Table 2). Plasmids pRIVM_C014947_5 and pRIVM_C014947_6 from KpnCluster-021 had 100% sequence coverage with 92.18 to 99.99% identity with plasmids isolated from Enterobacter hormaechei. Plasmids similar to the eight plasmids from KpnCluster-021 were detected previously in a variety of hosts, e.g. Salmonella enterica, K. pneumoniae, and E. hormaechei, suggesting these plasmids are broad-host range. The fact that 15 of the 22 plasmids analyzed in this study were found previously in distinct hosts, suggest international spread of these plasmids.
Table 2.
BLAST similarity analysis of K. pneumoniae plasmids.
| Plasmid | KpnCluster | blaKPC allele | Bacterial species | Plasmid | Query coverage (%) | Identity (%) | Accession number | Country | Year | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| pRIVM_C008981_1 | KpnCluster-003 | blaKPC-2 | K. pneumoniae | pGMI16-005_01 | 35 | 99.96 | CP028181.1 | Denmark | 2013 | |
| pRIVM_C014906_1 | KpnCluster-019 | blaKPC-2 | K. pneumoniae | pKPN1482-1 | 63 | 99.91 | CP020842.1 | USA | 2014 | Long et al. (2017)42 |
| pRIVM_C014906_2 | K. quasipneumoniae | plasmid pG747 | 97 | 99.84 | CP034137.1 | Nigeria | 2013 | |||
| pRIVM_C014906_3 | blaKPC-2 | K. pneumoniae | unnamed5 | 58 | 99.97 | CP033630.1 | Italy | 2013 | Roe et al. (2019)43 | |
| pRIVM_C018535_1 | blaKPC-2 | K. pneumoniae | pKPN1482-1 | 63 | 99.91 | CP020842.1 | USA | 2014 | Long et al. (2017)42 | |
| pRIVM_C018535_2 | blaKPC-2 | K. pneumoniae | unnamed5 | 58 | 99.97 | CP033630.1 | Italy | 2013 | Roe et al. (2019)43 | |
| pRIVM_C014947_1 | KpnCluster-021 | S. enterica | pSJO-60984 | 93 | 99.99 | CP025277.1 | USA | 2007 | ||
| pRIVM_C014947_2 | K. pneumoniae | unnamed3 | 90 | 99.98 | CP032170.1 | USA | 2015 | |||
| pRIVM_C014947_3 | blaKPC-2 | K. pneumoniae | pA1705-KPC | 93 | 99.97 | MH909348.1 | China | 2013 | ||
| pRIVM_C014947_4 | K. pneumoniae | pKP18-2079_5kb | 100 | 100 | MT090963.1 | China | 2018 | |||
| pRIVM_C014947_5 | E. hormaechei | pC4_003 | 100 | 99.97 | CP042543.1 | Australia | 2007 | |||
| pRIVM_C014947_6 | K. pneumoniae | pD17KP0032-3 | 100 | 100 | CP052331.1 | S. Korea | 2017 | |||
| pRIVM_C014947_7 | E. hormaechei | unnamed3 | 97 | 100 | CP035388.1 | UK | 2016 | |||
| pRIVM_C014947_8 | E. hormaechei | pC45-004 | 100 | 99.69 | CP042555.1 | Australia | 2013 | |||
| pRIVM_C015139_1 | KpnCluster-005 | K. pneumoniae | plasmid 2 | 100 | 99.99 | LR130549.1 | Australia | 2018 | ||
| pRIVM_C015139_2 | blaKPC-2 | K. pneumoniae | pUJ-83KPC | 99 | 98.55 | MG700549.1 | Germany | 2017 | ||
| pRIVM_C015139_3 | K. pneumoniae | pBK13043-2 | 100 | 99.89 | CP020839.1 | USA | 2004 | Long et al. (2017)42 | ||
| pRIVM_C015274_1 | KpnCluster-008 | blaKPC-3 | K. pneumoniae | plasmid p2 | 87 | 99.91 | CP019774.1 | Switzerland | 2015 | Ruppe et al. (2017)44 |
| pRIVM_C015274_2 | K. pneumoniae | pBK13043-2 | 100 | 99.95 | CP020839.1 | USA | 2004 | Long et al. (2017)42 | ||
| pRIVM_C015274_3 | K. pneumoniae | ColEST258 | 100 | 100 | JN247853.1 | Italy | 2012 | Garcia-Fernandez et al. (2012)45 | ||
| pRIVM_C015451_1 | KpnCluster-025 | blaKPC-3 | K. pneumoniae | pKPC | 99 | 99.99 | CP043971.1 | France | 2019 | |
| pRIVM_C015451_2 | K. pneumoniae | p911021-tetA | 66 | 99.55 | MG288679.1 | China | 2019 |
Prophage sequences in the K. pneumoniae cluster genomes
PHASTER analysis revealed that the majority of the large and medium-sized plasmids from different genetic clusters with IncFIB(K) or IncFIB(pQil) and IncFII(K) replicons contained one to four regions with prophage-related sequences e.g. genes encoding putative phage integrase, phage-like proteins, coat proteins, and/or tail shaft proteins (Table 3). The size of the prophage sequence regions varied per plasmid. The most commonly found prophage-related sequence in large and medium-sized plasmids of cluster isolates was an Escherichia phage RCS47 (Table 3). This sequence entails the 14.2 kb ygbMLKJI-blaSHV-recF-lacY region flanked by IS26 elements and representing 12% of the RCS47 prophage genome. The small plasmids of < 50 kb lacked phage-related sequences. In contrast, the chromosomes of cluster isolates carried at least three to nine phage sequence regions covering 10–50% of the phage genome. These phage sequence regions covered a wide variety of distinct phages, including prophage sequences from Salmonella, Klebsiella, Cronobacter, Enterobacteria phages (Suppl. Table 1). The most commonly found prophage sequence in Klebsiella chromosomes was the Enterobacteria phage P4.
Table 3.
Predicted prophage sequences among K. pneumoniae plasmids.
| Plasmid(s) | blaKPC allele | Phage region(s) length (kb) | Most common phage | Accession number |
|---|---|---|---|---|
| pRIVM_C008981_1 | blaKPC-2 | 11.3 | Escherichia phage RCS47 | NC_042128 |
| pRIVM_C014906_1 | blaKPC-2 | 7, 3.2, 23.3, 5.1, 5.7 | Stx2-converting phage 1717 | NC_011357 |
| pRIVM_C014906_2 | 43.6 | Escherichia phage RCS47 | NC_042128 | |
| pRIVM_C014906_3 | blaKPC-2 | |||
| pRIVM_C018535_1 | blaKPC-2 | 7, 3.2, 23.3, 5.1, 5.7 | Stx2-converting phage 1717 | NC_011357 |
| pRIVM_C018535_2 | blaKPC-2 | |||
| pRIVM_C014947_1 | 10.9 | Bacillus phage Shanette | NC_028983 | |
| pRIVM_C014947_2 | ||||
| pRIVM_C014947_3 | blaKPC-2 | |||
| pRIVM_C014947_4 | ||||
| pRIVM_C014947_5 | ||||
| pRIVM_C014947_6 | ||||
| pRIVM_C014947_7 | ||||
| pRIVM_C014947_8 | ||||
| pRIVM_C015139_1 | 23.5, 11.3 | Stx2-converting phage 1717, Escherichia phage RCS47 | NC_011357, NC_042128 | |
| pRIVM_C015139_2 | blaKPC-2 | 30.8 | Escherichia phage RCS47 | NC_042128 |
| pRIVM_C015139_3 | 24.8 | Escherichia phage RCS47 | NC_042128 | |
| pRIVM_C015274_1 | blaKPC-3 | 38.7 | Escherichia phage RCS47 | NC_042128 |
| pRIVM_C015274_2 | 24.8 | Escherichia phage RCS47 | NC_042128 | |
| pRIVM_C015274_3 | ||||
| pRIVM_C015451_1 | blaKPC-3 | 39.5 | Escherichia phage RCS47 | NC_042128 |
| pRIVM_C015451_2 | 20.6 | Staphylococcus phage SPbeta-like | NC_029119 |
Discussion
We showed that a K. pneumoniae ST560 strain carrying blaKPC-2 from KpnCluster-019 was transmitted between the Netherlands and the Caribbean. This is based on the high genetic relatedness of the 13 isolates from KpnCluster-019 as assessed by wgMLST and their highly similar resistome and plasmidome. We found that one person lived in the Caribbean and migrated to the Netherlands. After migration, a KpnCluster-019 isolate was obtained from this person in a Dutch hospital. Possibly other transmissions by other persons could have occurred, but these were not confirmed in this study. By combining short-read with long-read sequencing data, we identified 22 plasmids of seven K. pneumoniae isolates from six distinct genetic clusters found in the Netherlands and the Caribbean and analyzed these plasmids for its AMR gene profile, blaKPC transposons, replicons, transposon families, and gene content. The plasmid composition varied among the genetic clusters. Some of the cluster isolates had unique MLST sequence types (ST144 and ST560) which were not published previously and differ from globally circulating extensively drug-resistant (XDR) K. pneumoniae ST258 and ST307 strains23,24. KpnCluster-019 is unique compared to the other cluster isolates analyzed in this study for the following reasons. First, KpnCluster-019 harbors a unique and extensive set of AMR genes on the chromosome and in its plasmids. Secondly, KpnCluster-019 isolates were the only to contain three copies of the blaKPC-2 allele, two on two different plasmids and one in the chromosome. The localization of blaKPC-2 on the chromosome and additional blaKPC-2 copies have been reported previously and is further complicating the understanding of transmission of multidrug-resistant K. pneumoniae25,26. Thirdly, KpnCluster-019 and also KpnCluster-021 isolates from the Caribbean harbored the blaKPC-2 allele on a 5.6 kb ∆Tn4401a-like transposon, while the other isolates from the other genetic clusters from the Netherlands contained blaKPC on a 10 kb Tn4401a transposon. Most global descriptions of K. pneumoniae blaKPC the past decade have been associated with Tn4401a or isoforms hereof9. The traditional association of blaKPC with the Tn4401a transposon has possibly been eroded in K. pneumoniae isolates from the Caribbean to a smaller variant. This is the first report of identification of a 5.6 kb ∆Tn4401a-like blaKPC-2 transposon of K. pneumoniae in the Netherlands. Preliminary surveillance data analysis revealed that the ∆Tn4401a-like element carrying blaKPC-2 and smaller variants disseminated among E. cloacae, Serratia marcesens, K. oxytoca and E. coli in the Netherlands (unpublished data). Future work will seek to understand the dissemination of the ∆Tn4401a-like blaKPC-2 element among CPE in the Netherlands. Lastly, the plasmids of KpnCluster-019 isolates contained also unique plasmid content, including a distinct transposon signature, two toxin-antitoxin systems and a ceaC colicin which possibly contribute to the success in survival, niche adaptation or transmission of this strain.
The K. pneumoniae blaKPC-3 isolates had higher MICs for meropenem than the K. pneumoniae blaKPC-2 isolates, which is in line with a previous study21. The KPC-2 enzyme differs in a single amino acid substitution (Histidine 272 to Tyrosine) from KPC-3. Additional changes in KPC-3 can lead to increased resistance for ceftazidime and cephamycin27. The increase in meropenem resistance observed in our study is possibly correlated with improved ability of KPC-3 enzymes to hydrolyze the meropenem antibiotic15. Alternatively, additional beta-lactamase genes such as blaOXA-1, blaOXA-9 or blaTEM-1A may contribute to increased resistance for meropenem28.
Despite the limited number of long-read sequenced isolates, we have highlighted important new insights in the genomic surveillance of a notorious multi-antibiotic resistant nosocomial pathogen. In some clusters, the plasmidome varied as this was likely due to loss of a plasmid. Also, the resistome data suggest the presence of other plasmids in cluster isolates that were not present in the isolates that were sequenced using TGS. To overcome this limitation, all isolates used in this study should have been sequenced using long-read third generation sequencing. Nevertheless, we identified plasmids in K. pneumoniae blaKPC-2 and blaKPC-3 cluster isolates which vary in size from large, medium and small. The large and medium sized plasmids were enriched for a variety of transposons, conjugation transfer systems, cation efflux systems including Fe(3+)-dicitrate transport, and genes encoding for silver, copper and arsenic resistance. The small plasmids contained putative virulence determinants. The presence of these systems may contribute to the success of transmission of specific K. pneumoniae strains in the hospital setting or the community13,29,30. Escherichia RCS47 prophage sequences were found on medium and large plasmids in the cluster isolates analyzed. In contrast, the chromosomes contained a variety of prophage-related sequences. RCS47 is a P1-like bacteriophage carrying the ESBL-encoding blaSHV-2 gene was isolated from a clinical E. coli strain31. The prevalence of P1-like prophages in animal and human E. coli strain collections was 12.6%31. The presence of P1-like phage sequences in plasmids of a snapshot of the K. pneumoniae population in the Netherlands and the Caribbean suggest that the role of P1-like phages in disseminating antibiotic resistance may be underestimated32.
In conclusion, long-read sequencing contributed to the understanding of the successful transmission of the KpnCluster-019 K. pneumoniae blaKPC-2 strain. Plasmid content such as conjugation machinery, transposons, virulence determinants and phages may contribute to diversification, and dissemination of plasmids containing AMR genes, and therefore represent important plasmid features that warrants future investigation. More long-read plasmid sequencing efforts of CPE and K. pneumoniae in particular are required to identify the complete plasmid reservoir involved in the spread of antibiotic resistance determinants in the Netherlands and the Caribbean islands.
Methods
Bacterial isolates
For the Dutch National carbapenemase-producing Enterobacterales (CPE) Surveillance program, medical microbiology laboratories from the Netherlands and the Caribbean routinely send CPE isolates with a meropenem minimum inhibitory concentration (MIC) of ≥ 0.25 µg/ml and/or an imipenem MIC of ≥ 1 µg/ml or phenotypic (CIM-test) or genotypical evidence of carbapenemase production to the National Institute of Public Health and the Environment (RIVM)16. For this study, 84 carbapenemase-producing K. pneumoniae isolates carrying either the blaKPC-2 allele or the blaKPC-3 allele were included and collected in the period from January 1st 2014 until June 30th 2019. Only the first K. pneumoniae isolate per person in this study period was selected. The 84 isolates were obtained from 84 persons and from various isolation sites, i.e. rectum/perineum (n = 43), throat (n = 11), pus (n = 2), sputum (n = 4), urine (n = 10), wound (n = 5) and nine were from miscellaneous isolation sites. All bacterial strains were grown aerobically at 37 °C on Columbia sheep blood agar plates.
Antimicrobial susceptibility testing
Resistance to carbapenem was confirmed by assessing the MICs for meropenem for all the 84 isolates using an Etest (bioMérieux Inc., Marcy l’Etoile, France). Based on the clinical breakpoints according to EUCAST, the K. pneumoniae isolates were classified as sensitive (≤ 2 mg/L), intermediate (> 2 mg/L and ≤ 8 mg/L) and resistant (> 8 mg/L) to meropenem. In addition, all isolates were analyzed for the production of carbapenemase using the carbapenem inactivation method (CIM) as described previously33.
Next-generation sequencing, MLST and wgMLST
All 84 K. pneumoniae isolates were subjected to next-generation sequencing (NGS) using the Illumina HiSeq 2500 (BaseClear, Leiden, the Netherlands). The NGS data of the K. pneumoniae isolates were used for classical MLST and wgMLST analyses using the in-house wgMLST scheme in SeqSphere software version 6.0.2 (Ridom GmbH, Münster, Germany). The in-house K. pneumoniae wgMLST scheme was comprised of 4978 genes (3471 core-genome and 1507 accessory-genome targets) using K. pneumoniae MGH 78,578 (NC_009648.1) as a reference genome21. For classical MLST, the existing scheme was used and cluster nomenclature were depicted in Table 134. The resulting data was imported into Bionumerics version 7.6.3 for subsequent comparative analyses (Applied Maths, Sint-Martens-Latem, Belgium). The antibiotic resistance gene profile and plasmid replicon compositions in all of the isolates were determined by interrogating the online ResFinder (version 3.1.0) and PlasmidFinder (version 2.0.2) databases available at the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/services/)35,36. For ResFinder, a 90% identity threshold and a minimum length of 60% were used as criteria, whereas for PlasmidFinder, an identity of 95% was utilized.
Long-read third-generation sequencing
One K. pneumoniae isolate per genetic KpnCluster was sequenced using long-read third-generation Nanopore sequencing18,37. High molecular weight DNA was isolated using an in-house developed protocol. Bacteria were grown overnight in 1.5 ml Brain heart infusion broth and culture was spun down at 13,000 × g for 2 min. The pellet was washed and resuspended in 500 µl of 150 mM NaCl. The suspension was spun down at 5000 × g for 5 min and the pellet was resuspended in 100 µl of QuickExtract DNA Extraction Solution (Lucigen) and 0.1 µl Ready-Lyse Lysozyme solution (Epicentre) and incubated for 1 h at 37 °C. Subsequently, 85 µl 10 mM Tris 1 mM EDTA pH = 8 (1 × TE), 10 µl proteinase K (> 600 mAU/mL, Qiagen) and 5 µl 20% sodium dodecyl sulfate solution were added, and the mixture was incubated at 56 °C for 30 min. DNA was precipitated overnight at -20 °C by adding 0.1 × volume 3 M sodium acetate pH = 5.2 and 2.5 × volume ice cold 100% ethanol. Precipitated DNA was spun down at 13,000 × g for 15 min and pellets were washed with 1 ml 70% ethanol followed by another centrifugation at 13,000 × g for 5 min. After drying, the pellet was dissolved in 200 µl 1 × TE and diluted to 1 µg with Nuclease-free water.
The Oxford Nanopore protocol SQK-LSK108 (https://community.nanoporetech.com) and the expansion kit for native barcoding EXP-NBD104 was used. Briefly, a shearing step was performed using g-TUBE’s (Covaris) to obtain an average DNA fragment size of 8 kb. The DNA was repaired using FFPE and end-repair kits (New England BioLabs) followed by ligation of barcodes with bead clean up using AMPure XP (Beckman Coulter) after each step. Barcoded isolates were pooled and sequencing adapters were added by ligation. The final library was loaded onto a MinION flow cell (MIN-106 R9.4.1). The 48-h sequence run was started without live base calling enabled on a MinION device connected to a desktop computer. After the sequence run, base calling and de-multiplexing was performed using Albacore 2.3.1 and a single FASTA file per isolate was extracted from the FAST5 files using Poretools 0.5.138. Illumina and Nanopore data were used in a hybrid assembly performed by Unicycler v0.4.439. The resulting contig files were annotated using Prokka and were subsequently loaded into BioNumerics for further analyses40.
Minimum spanning tree and UPGMA analyses
The BioNumerics software was used to generate a minimum spanning tree (MST) or an UPGMA hierarchical clustering as described previously16. The MST was based on an in-house K. pneumoniae wgMLST scheme. The categorical coefficient was used to calculate the MST. wgMLST clusters were defined as a minimum of two isolates of which the genetic distance between the two isolates was ≤ 20 genes (20/4978 ≤ 0.4% different). An UPGMA clustering of K. pneumoniae blaKPC-2 and blaKPC-3 isolates was performed based on the presence and/or absence of antibiotic resistance genes per isolate.
Plasmid reconstruction by read mapping
The CLC Genomics Workbench version 12.0 software (www.qiagenbioinformatics.com) was used to reconstruct plasmids. For this, complete plasmids obtained by TGS were used as a scaffold to map the trimmed NGS reads of isolates that were from the same genetic wgMLST cluster. A plasmid was scored “present” in an isolate if reads mapped to a reference plasmid of interest and ≥ 85% of the consensus sequence size in kilo bases was reconstructed. Linear DNA fragments < 5 kb were omitted in this study. Nucleotide BLAST analyses on plasmid sequences were performed using the https://blast.ncbi.nlm.nih.gov website and date from October 2019.
Plasmid content analysis
Bionumerics was used to extract and analyze annotated genes and tranposases in the 22 different plasmids. The data was plotted in Excel. Phaster, the PHAge Search Tool Enhanced Release website (https://phaster.ca/) was used to determine the presence of phage sequences in the plasmids and searches date from October 201941.
Ethics statement
The bacterial isolates used in this study belong to the medical microbiological laboratories participating in the Dutch National CPE Surveillance program and was obtained as part of routine clinical care in the past. Since no identifiable personal data were collected and data were analyzed anonymously, written or verbal patient consent was not required for this study and was therefore not obtained. According to the Dutch Medical Research Involving Human Subjects Act (WMO) this study was considered exempt from review by an Institutional Review Board.
Supplementary information
Acknowledgements
We thank all the members of the Dutch CPE surveillance study Group and the Dutch medical microbiology laboratories for submitting CPE isolates to the RIVM for the national CPE surveillance program. We thank Dr. Judith W.A. Hoogenboom-Beuving for searching for an epidemiological link of KpnCluster-019 isolates from the Netherlands with the Caribbean. We also thank Prof. Dr. E. Kuijper, Dr. M.G. Mennen and Dr. D.W. Notermans for critical reading of this manuscript.
Author contributions
A.P.A.H. and L.M.S. were involved in the experimental design, coordinated the whole work, summarized the data, prepared the figures and wrote the manuscript. FL, ADH and MGSV performed laboratory experiments. F.L., D.B., S.W., M.G.S.V., and H.V.D.H. processed sequencing data and curated databases. D.B. and H.V.D.H. were involved in bioinformatic analyses. Members of the Dutch CPE surveillance Study Group provided bacterial isolates used in this study. All authors and contributors reviewed the manuscript.
Data availability
The Illumina (NGS) and plasmid sequence data sets generated and analyzed in this study are freely available in the Sequence Read Archive (SRA) under BioProject ID PRJNA634885 and in Genbank under accession numbers as depicted in Supplementary Table 2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A comprehensive list of consortium members appears at the end of the paper.
Contributor Information
Antoni P. A. Hendrickx, Email: antoni.hendrickx@rivm.nl
The Dutch CPE surveillance Study Group:
T. Halaby, R. Steingrover, J. W. T. Cohen Stuart, D. C. Melles, K. van Dijk, I. J. B. Spijkerman, D. W. Notermans, J. H. Oudbier, M. L. van Ogtrop, A. van Dam, M. den Reijer, J. A. J. W. Kluytmans, M. P. M. van der Linden, E. E. Mattsson, M. van der Vusse, E. de Jong, A. Maijer-Reuwer, M. van Trijp, A. J. van Griethuysen, A. Ott, E. Bathoorn, J. C. Sinnige, E. Heikens, E. I. G. B. de Brauwer, F. S. Stals, W. Silvis, J. W. Dorigo-Zetsma, K. Waar, S. P. van Mens, N. Roescher, A. Voss, H. Wertheim, B. C. G. C. Slingerland, H. M. E. Frenay, T. Schulin, B. M. W. Diederen, L. Bode, M. van Rijn, S. Dinant, M. Damen, P. de Man, M. A. Leversteijn-van Hall, E. P. M. van Elzakker, A. E. Muller, P. Schneeberger, D. W. van Dam, A. G. M. Buiting, A. L. M. Vlek, A. Stam, A. Troelstra, I. T. M. A. Overdevest, R. W. Bosboom, T. A. M. Trienekens, M. J. H. M. Wolfhagen, and S. Paltansing
Supplementary information
is available for this paper at 10.1038/s41598-020-73440-2.
References
- 1.van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. N. Y. Acad. Sci. 2019 doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 3.Bonomo RA, et al. Carbapenemase-producing organisms: a global scourge. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018;66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trecarichi EM, Tumbarello M. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence. 2017;8:470–484. doi: 10.1080/21505594.2017.1292196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018;45:131–139. doi: 10.1016/j.mib.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 7.Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Rozwandowicz M, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 9.Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob. Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas T, et al. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz-Price LS, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambler RP, et al. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathers AJ, Peirano G, Pitout JDD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walther-Rasmussen J, Høiby N. Class A carbapenemases. J. Antimicrob. Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 15.Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 2005;49:4760–4762. doi: 10.1128/AAC.49.11.4760-4762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch T, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae in a Dutch Hospital, with interspecies transfer of the resistance plasmid and unexpected occurrence in unrelated health care centers. J. Clin. Microbiol. 2017;55:2380–2390. doi: 10.1128/JCM.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019;20:356–370. doi: 10.1038/s41576-019-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, Giordano F, Ning Z. Oxford nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 2016;14:265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lannoy C, de Ridder D, Risse J. The long reads ahead: de novo genome assembly using theMiniON. F1000Research. 2017;6:1083. doi: 10.12688/f1000research.12012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Centre for Disease Prevention and Control. Carbapenemase-producing bacteria in Europe: interim results from the European Survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE) project. (2013).
- 21.van der Zwaluw K, et al. Molecular characteristics of carbapenemase-producing Enterobacterales in the Netherlands; results of the 2014–2018 national laboratory surveillance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020 doi: 10.1016/j.cmi.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Liang W, et al. KlcAHS genes are ubiquitous in clinical, blaKPC-2-positive, Klebsiella pneumoniae isolates. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019;70:84–89. doi: 10.1016/j.meegid.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Holt KE, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3574–3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019 doi: 10.1128/CMR.0001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, et al. Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M Integrated in the Chromosome. Antimicrob. Agents Chemother. 2017 doi: 10.1128/AAC.00076-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Y, Liu L, McNally A, Zong Z. Coexistence of three blaKPC-2 genes on an IncF/IncR plasmid in ST11 Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2019;17:90–93. doi: 10.1016/j.jgar.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Haidar G, et al. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 2017 doi: 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocsis E, et al. blaNDM-1 carriage on IncR plasmid in Enterobacteriaceae strains. Microb. Drug Resist. 2016;22:123–128. doi: 10.1089/mdr.2015.0083. [DOI] [PubMed] [Google Scholar]
- 29.Wyres KL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanczvikkel A, Füzi M, Ungvári E, Tóth Á. Transmissible silver resistance readily evolves in high-risk clone isolates of Klebsiella pneumoniae. Acta Microbiol. Immunol. Hung. 2018;65:387–403. doi: 10.1556/030.65.2018.031. [DOI] [PubMed] [Google Scholar]
- 31.Billard-Pomares T, et al. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 2014;58:6550–6557. doi: 10.1128/AAC.03183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venturini C, et al. Diversity of P1 phage-like elements in multidrug resistant Escherichia coli. Sci. Rep. 2019;9:18861. doi: 10.1038/s41598-019-54895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Zwaluw K, et al. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE. 2015;10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loose M, Malla S, Stout M. Real-time selective sequencing using nanopore technology. Nat. Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loman NJ, Quinlan AR. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics. 2014;30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 41.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesley Long SW, et al. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: Unexpected abundance of clonal group 307. mBio. 2017;8(3):e00489-17. doi: 10.1128/mBio.00489-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roe CC, et al. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front. Microbiol. 2019;10:542. doi: 10.3389/fmicb.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruppé E, et al. Clonal or not clonal? Investigating hospital outbreaks of KPC-producing Klebsiella pneumoniae with whole-genome sequencing. Clin. Microbiol. Infect. 2017;237(7):470–475. doi: 10.1016/j.cmi.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 45.García-Fernández A, et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 2012;56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina (NGS) and plasmid sequence data sets generated and analyzed in this study are freely available in the Sequence Read Archive (SRA) under BioProject ID PRJNA634885 and in Genbank under accession numbers as depicted in Supplementary Table 2.