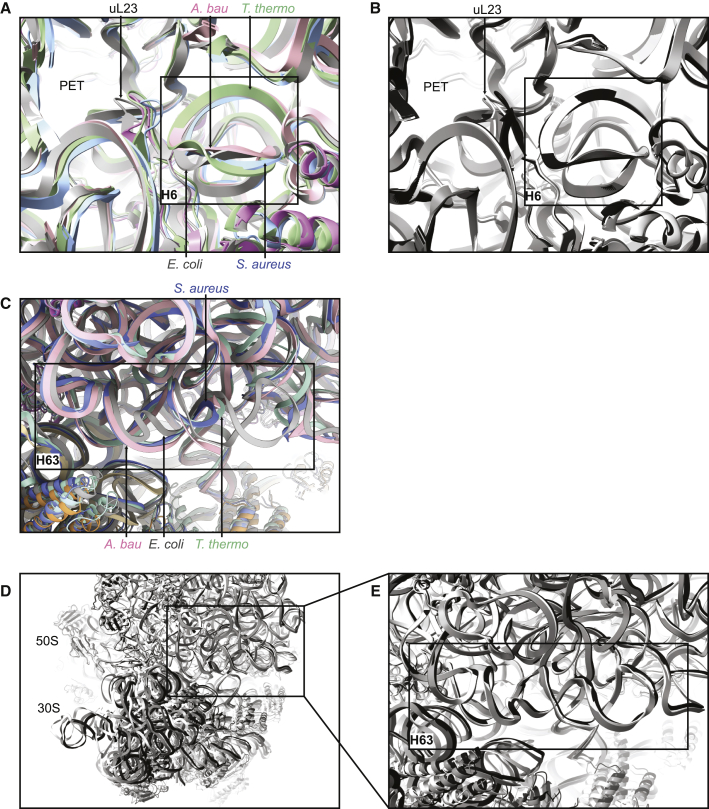
Figure 3.
Unique Structural Features in the 23S rRNA Helices H6 and H63
(A) H6 of A. baumannii (pink) takes a different conformation compared with H6 of E. coli (gray, PDB: 4YBB), S. aureus (blue, PDB: 5LI0), and T. thermophilus (green, PDB: 5E81).
(B) H6 maintains the same conformation across various E. coli ribosome structures, as shown here in a crystal structure (light gray, PDB: 4YBB ribosome I; mid-gray, PDB: 4YBB ribosome II) and an EM structure (dark gray, PDB: 5MDZ), unlike the nearby β-hairpin loop of uL23, which shows some variation.
(C) H63 of A. baumannii (pink) takes a different conformation than H63 of E. coli (gray, PDB: 4YBB) and is longer than in S. aureus (blue, PDB: 5LI0) and T. thermophilus (green, PDB: 5E81).
(D) E. coli ribosome models aligned on the 50S ribosomal subunit, representing a range of rotation states of the 30S ribosomal subunit. Empty ribosome in an intermediate rotated state (light gray, PDB: 4YBB ribosome I), empty ribosome in non-rotated state (mid-gray, PDB: 4YBB ribosome II), and ribosome with A-site and P-site tRNA (dark gray, PDB: 5MDZ).
(E) H63 in these three E. coli ribosome models. Despite the difference in intersubunit rotation states, the conformation of H63 remains similar across the three models.
The structure of the A. baumannii ribosome-amikacin complex is used, but the structures of all highlighted regions hold true for the A. baumannii ribosome-tigecycline complex. PET, polypeptide exit tunnel.