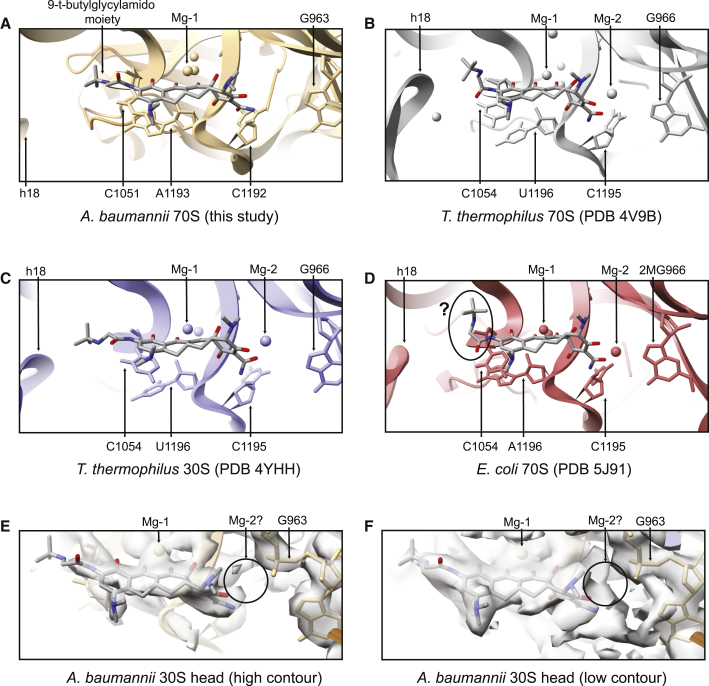
Figure 6.
The Primary Tigecycline Binding Site in Ribosomes and Ribosome Subunits of Different Bacteria
(A–D) Atomic models of the primary tigecycline binding site in ribosomes and ribosome subunits of different bacteria. The nature of tigecycline binding is broadly similar across the structures, with some differences in the stacking interaction of tigecycline with C1051 (C1054) of 16S rRNA, the conformation of the 9-t-butylglycylamido moiety, and the coordination of a second magnesium ion. (A) Atomic model of tigecycline (gray) bound to the 70S of the A. baumannii ribosome (brown). The 30S head model and h18 from the 30S body model are shown. The consensus map was used to confirm the relative proximity of these features. (B) Atomic model of tigecycline (gray) bound to the 70S T. thermophilus ribosome (gray, PDB: 4V9B). (C) Atomic model of tigecycline (gray) bound to the 30S T. thermophilus ribosomal subunit (blue, PDB: 4YHH). (D) Atomic model of tigecycline (gray) bound to the 70S E. coli ribosome (red, PDB: 5J91). The density is not strong enough to support either an extended or a bent conformation of the 9-t-butylglycylamido moiety, as indicated by the question mark.
(E) EM density of the 30S head of the A. baumannii ribosome-tigecycline complex at the primary tigecycline site, high-contour level.
(F) EM density of the 30S head of the A. baumannii ribosome-tigecycline complex at the primary tigecycline site, low-contour level. It is difficult to discern possible magnesium ion density (Mg-2) from noise.