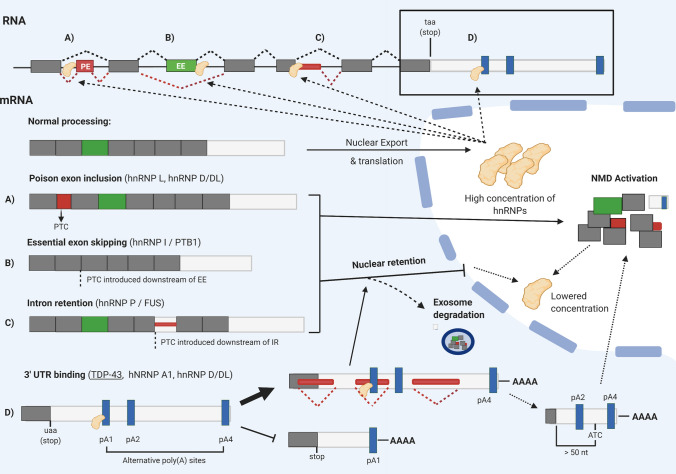
Fig. 4.
HnRNP autoregulation mechanisms. HnRNPs autoregulate their expression by several RNA processing mechanisms. HnRNP binding promotes specific splicing events that result in the production of NMD-sensitive mRNAs and/or transcripts confined to the nucleus (blue background). These include the activation of a normally skipped premature termination codon (PTC)-containing ‘poison exon’ (a), the skipping of a normally ‘essential exon’ (EE) (b) or retention of intronic RNA (IR) (c). TDP-43 binds to its 3′UTR TARDBP binding site within intron 7 and inhibits the selection of the proximal poly(A) site (pA1), up-regulating alternative polyadenylation at its more distal sites: pA4 and more rarely pA2 (isoform not shown) (d). The unstable isoform generated is detained in the nucleus and is subject to exosome-mediated degradation. TDP-43-binding and subsequent RNA Pol II stalling can also lead to alternative splicing of 3′ UTR intronic regions (red rectangles) which truncates the final exon, eliminates the true stop signal and exposes an alternative termination codon (ATC). The ATC being > 50 nt from the final exon-junction complex designates the transcript for NMD. This splicing event is not believed to significantly contribute to TDP-43 autoregulation, but is a crucial feature of hnRNP A1 and hnRNP D/DL autoregulatory mechanisms which activate 3′ UTR poison exon/intron events