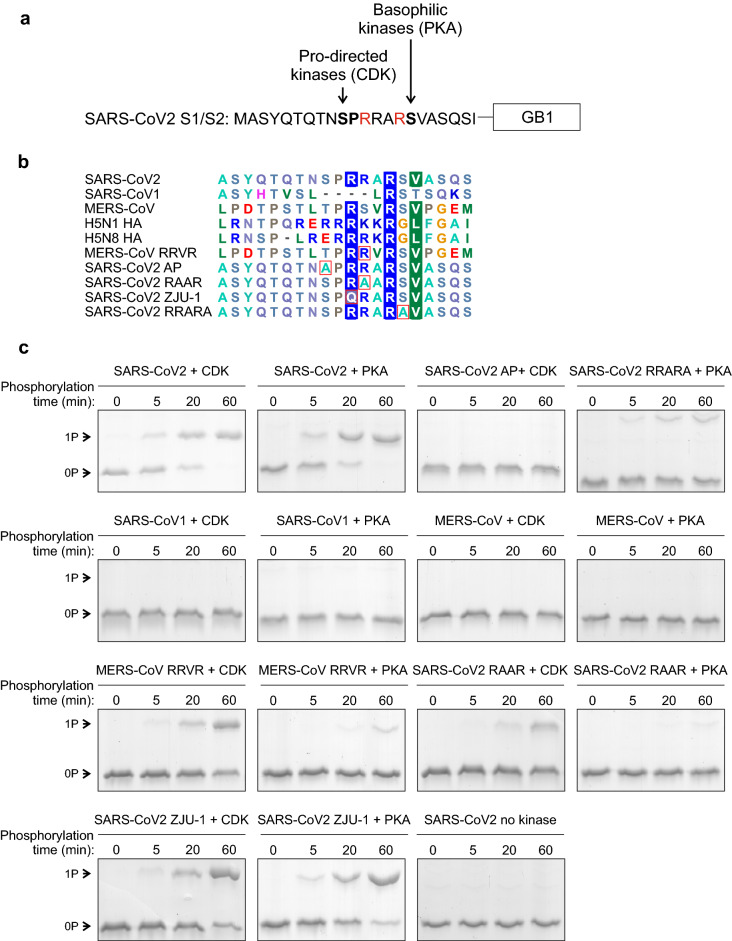
Figure 3.
Potential phosphorylation of the S1/S2 site. (a) A scheme of the 20-residue segment of the SARS-CoV2 S1/S2 site. In addition to a furin site, the four-residue insertion (PRRA) creates potential phosphorylation sites for proline-directed kinases (S680) and basophilic kinases (S686). (b) Multiple sequence alignment of different S1/S2 sequences used in phosphorylation assays in panel ‘c’. (c) The phosphorylation of S1/S2-GB1 reporter proteins was analyzed in vitro using cyclin B-Cdk1 (CDK) and protein kinase A (PKA) as a representative of proline-directed and basophilic kinases, respectively. The phosphorylation reactions were stopped at 0, 5, 20, and 60 min. Phosphorylation shifts were analyzed using Phos-tag SDS-PAGE, which separates the phosphorylated form from the unphosphorylated form. Coomassie-stained gels are shown. Uncropped images are shown in Supplementary Fig. S3.