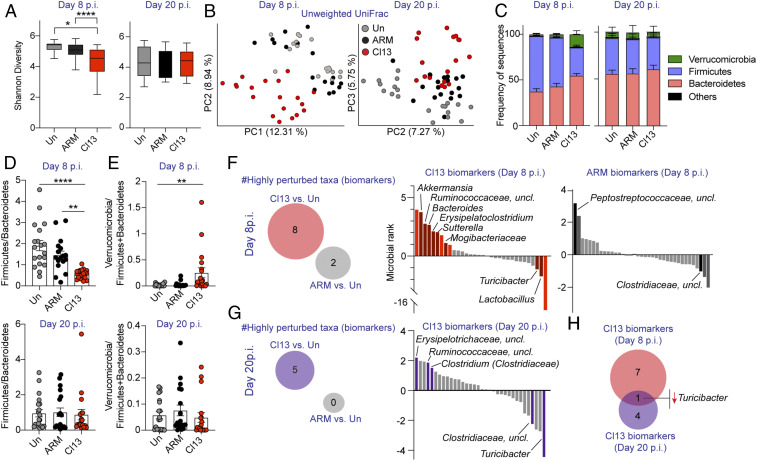
Fig. 1.
LCMV Cl13 infection induced intestinal dysbiosis that was mostly transient, uncoupled from viral loads, and characterized by reduced Firmicutes-to-Bacteroidetes ratio and increased Verrucomicrobia. See also SI Appendix, Fig. S1 and Datasets S1–S4. C57BL/6 mice were infected with LCMV ARM, Cl13, or left uninfected (Un) and 16S rRNA gene amplicon sequencing was performed on colonic and caecal contents from days 8 and 20 (p.i.). (A) Alpha-diversity by the Shannon diversity index at indicated time points. (B) Beta-diversity PCoA with unweighted Unifrac distance at indicated time points. PC1 was omitted for PCoA from the day 20 p.i. time point as it captured the basal differences of the cohorts used in the two experiments, rather than infection type. (C) Frequency of sequences at the phylum level at indicated time points. (D and E) Frequency of phylum Firmicutes divided by Bacteroidetes (D) or Verrucomicrobia divided by Firmicutes and Bacteroidetes (E) for each individual mouse at indicated time points. (F and G) Songbird MR analysis of genera in mice infected with ARM or Cl13 vs. Un on day 8 p.i. (F) or day 20 p.i. (G). Taxa with microbial ranks higher than 1 or lower than −1 in two independent repeats are considered highly perturbed (i.e., biomarkers). (Left) Total number of genera highly perturbed by Cl13 or ARM infections. (Right) The x axes correspond to individual taxa, taxa highly perturbed are indicated by text and colored in red (F) or violet (G) for Cl13 infection and black (F) for ARM infection. (H) Venn diagram overlapping biomarkers identified in F and G for day 8 (red) and day 20 (violet) after Cl13 infection. (A, C, and D) Graphs show averages plus min and max (A) or ± SEM (C–E). (A–E) Data are represented by pooling n = 8 to 10 mice/group from two independent experiments. (F–H) Data are representative of two independent experimental repeats. (A) Pairwise Kruskal–Wallis with false discovery rate (FDR) (Benjamini–Hochberg) correction. (D and E) Kruskal–Wallis with Dunn’s correction; *p-val < 0.05, **<0.01, ****<0.0001.